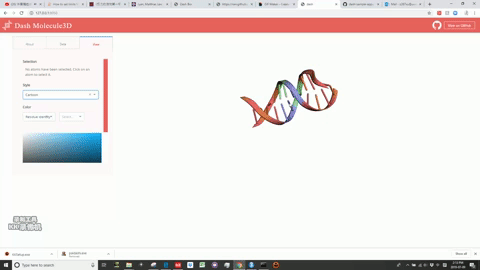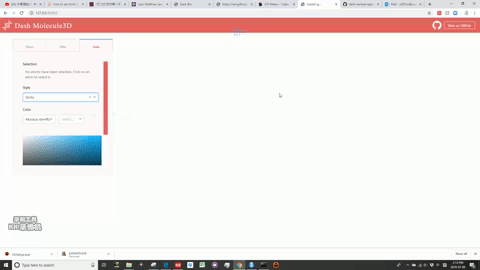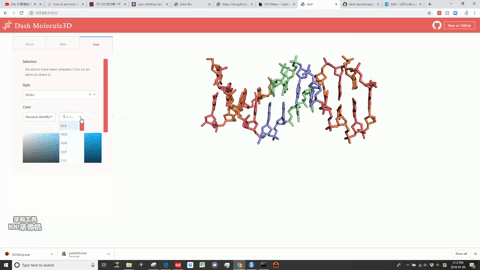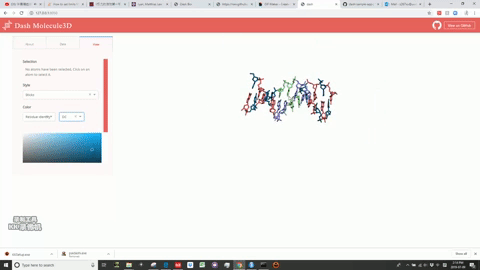
This is a Dash for R version of the molecule3d app previously written in Python.
Molecule3D is a visualizer that allows you to view biomolecules in multiple representations: sticks, spheres, and cartoons. "pdb" files are required for the database.
Clone the git repo and cd to the root directory:
git clone https://github.com/plotly/dash-sample-apps
cd dash-sample-apps/apps/dashr-molecule3d
Launch R, then install the required package dependencies in the default location:
remotes::install_github("plotly/dash-html-components")
remotes::install_github("plotly/dash-core-components")
remotes::install_github("plotly/dash-table")
remotes::install_github("plotly/dashR")
remotes::install_github("plotly/dash-daq")
remotes::install_github("plotly/dash-bio")
install.packages("bio3d")
install.packages("jsonlite")
Run the app, either interactively in R or RStudio, or by using the command line:
Rscript app.R
Finally, open your browser and enter the URL http://127.0.0.1:8050.