contrastyou is a scalable medical image segmentation framework realized during my Ph. D studies at
the École de technologie supérieure (ÉTS).
This framework resulted in several papers, including:
-
Boundary-aware Information Maximization for Self-supervised Medical Image Segmentation
-
Diversified Multi-prototype Representation for Semi-supervised Segmentation
-
Self-Paced Contrastive Learning for Semi-supervised Medical Image Segmentation with Meta-labels
-
Boosting semi-supervised image segmentation with global and local mutual information regularization
-
Mutual information deep regularization for semi-supervised segmentation
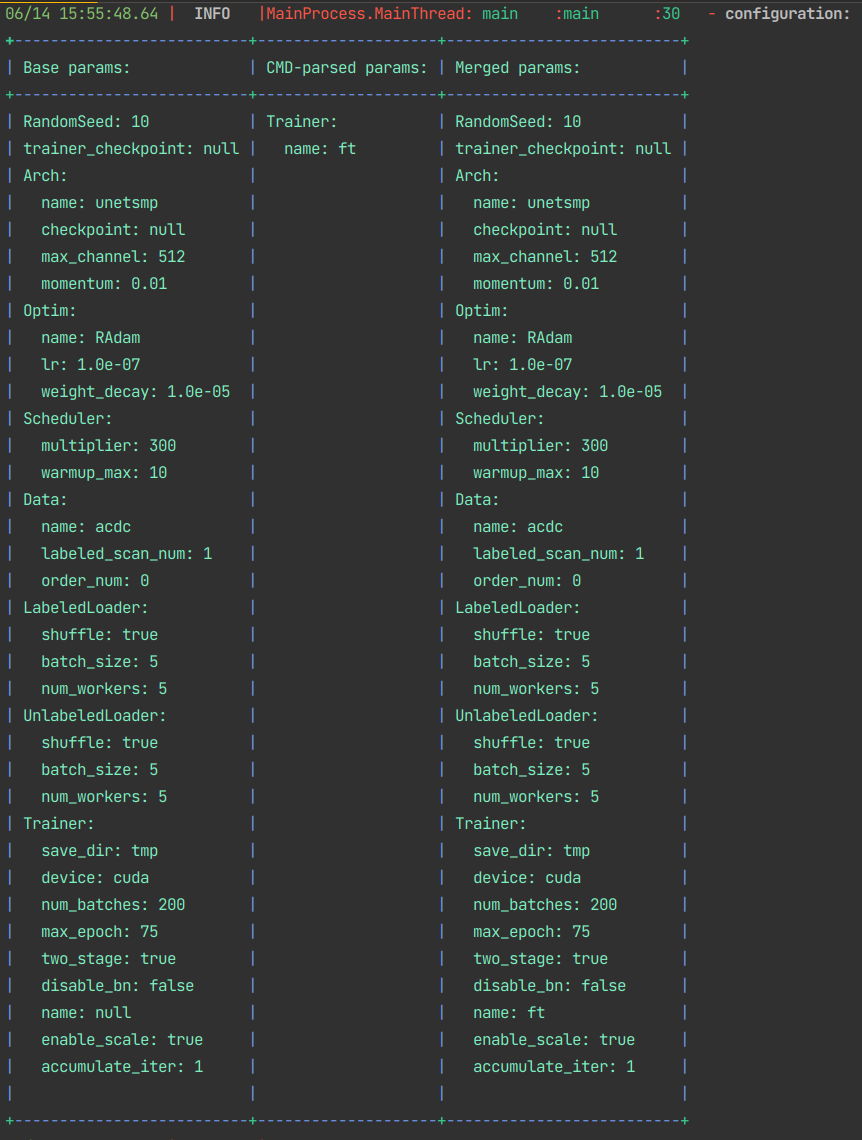
- Configurable configuration management
python main_nd.py -o Trainer.name=ft -p config/base.yaml- Automatic Trainer checkpointing and resuming
class Trainer(IOMixin, HookMixin, DDPMixin, AMPScalerMixin, ModuleBase):
RUN_PATH = MODEL_PATH # type:str # absolute path
def __init__(self, *, model: nn.Module, criterion: LossClass[Tensor], tra_loader: SizedIterable,
val_loader: SizedIterable, save_dir: str, max_epoch: int = 100, num_batches: int = 100, device="cpu",
config: Dict[str, Any], enable_scale: bool = False, accumulate_iter: int = 1) -> None:
super().__init__(enable_scale=enable_scale, accumulate_iter=accumulate_iter)
self._initialized = False
self._model = self._inference_model = model
self._criterion = criterion
self._tra_loader = tra_loader
self._val_loader = val_loader
self._save_dir = cast(str, Buffer(save_dir)) # persisting automatically
self._max_epoch = cast(int, Buffer(max_epoch)) # persisting automatically
self._num_batches = Buffer(num_batches) # persisting automatically
self._device = device
self.config = cast(dict, Buffer(config)) # persisting automatically
self._config = self.config- Extenable Epochers
class EpocherBase(AMPScaler, DDPMixin, metaclass=ABCMeta):
""" EpocherBase class to control the behavior of the training within one epoch.
>>> hooks = ...
>>> epocher = EpocherBase(...)
>>> with epocher.register_hook(*hooks):
>>> epocher.run()
>>> epocher_result, best_score = epocher.get_metric(), epocher.get_score()
"""
...
class SemiSupervisedEpocher(EpocherBase, ABC):
meter_focus = "semi"
def _assertion(self):
assert_transform_freedom(self._labeled_loader, False)
if self._unlabeled_loader is not None:
assert_transform_freedom(self._unlabeled_loader, False)
def __init__(self, *, model: nn.Module, optimizer: optimizerType, labeled_loader: SizedIterable,
unlabeled_loader: SizedIterable, sup_criterion: criterionType, num_batches: int, cur_epoch=0,
device="cpu", two_stage: bool = False, disable_bn: bool = False, scaler: GradScaler,
accumulate_iter: int = 1, **kwargs) -> None:
super().__init__(model=model, num_batches=num_batches, cur_epoch=cur_epoch, device=device, scaler=scaler,
accumulate_iter=accumulate_iter)
self._optimizer = optimizer
self._labeled_loader: SizedIterable = labeled_loader
self._unlabeled_loader: SizedIterable = unlabeled_loader
self._sup_criterion = sup_criterion
...
class FineTuneEpocher(SemiSupervisedEpocher, ABC):
meter_focus = "ft"
def __init__(self, *, model: nn.Module, optimizer: optimizerType, labeled_loader: dataIterType,
sup_criterion: criterionType, num_batches: int, cur_epoch=0, device="cpu", scaler: GradScaler,
accumulate_iter: int, **kwargs) -> None:
super().__init__(model=model, optimizer=optimizer, labeled_loader=labeled_loader, sup_criterion=sup_criterion,
num_batches=num_batches, cur_epoch=cur_epoch, device=device, scaler=scaler,
accumulate_iter=accumulate_iter, **kwargs)
def configure_meters(self, meters: MeterInterface) -> MeterInterface:
meters = super().configure_meters(meters)
meters.delete_meter("reg_loss")
return meters
...- Scalable Meters
class EvalEpocher(EpocherBase):
meter_focus = "eval"
...
def configure_meters(self, meters: MeterInterface) -> MeterInterface:
C = self.num_classes
report_axis = list(range(1, C))
meters.register_meter("loss", AverageValueMeter())
meters.register_meter("dice", UniversalDice(C, report_axis=report_axis))
return meters
...
def _batch_update(self, *, eval_img, eval_target, eval_group, file_names):
with self.autocast:
eval_logits = self._model(eval_img)
onehot_target = class2one_hot(eval_target.squeeze(1), self.num_classes)
eval_loss = self._sup_criterion(eval_logits.softmax(1), onehot_target, disable_assert=True)
self.meters["loss"].add(eval_loss.item())
self.meters["dice"].add(eval_logits.max(1)[1], eval_target.squeeze(1), group_name=eval_group)
....- Confortable logging with Loguru
 logger with info:
logger with info: Export LOGURU_LEVEL=INFO
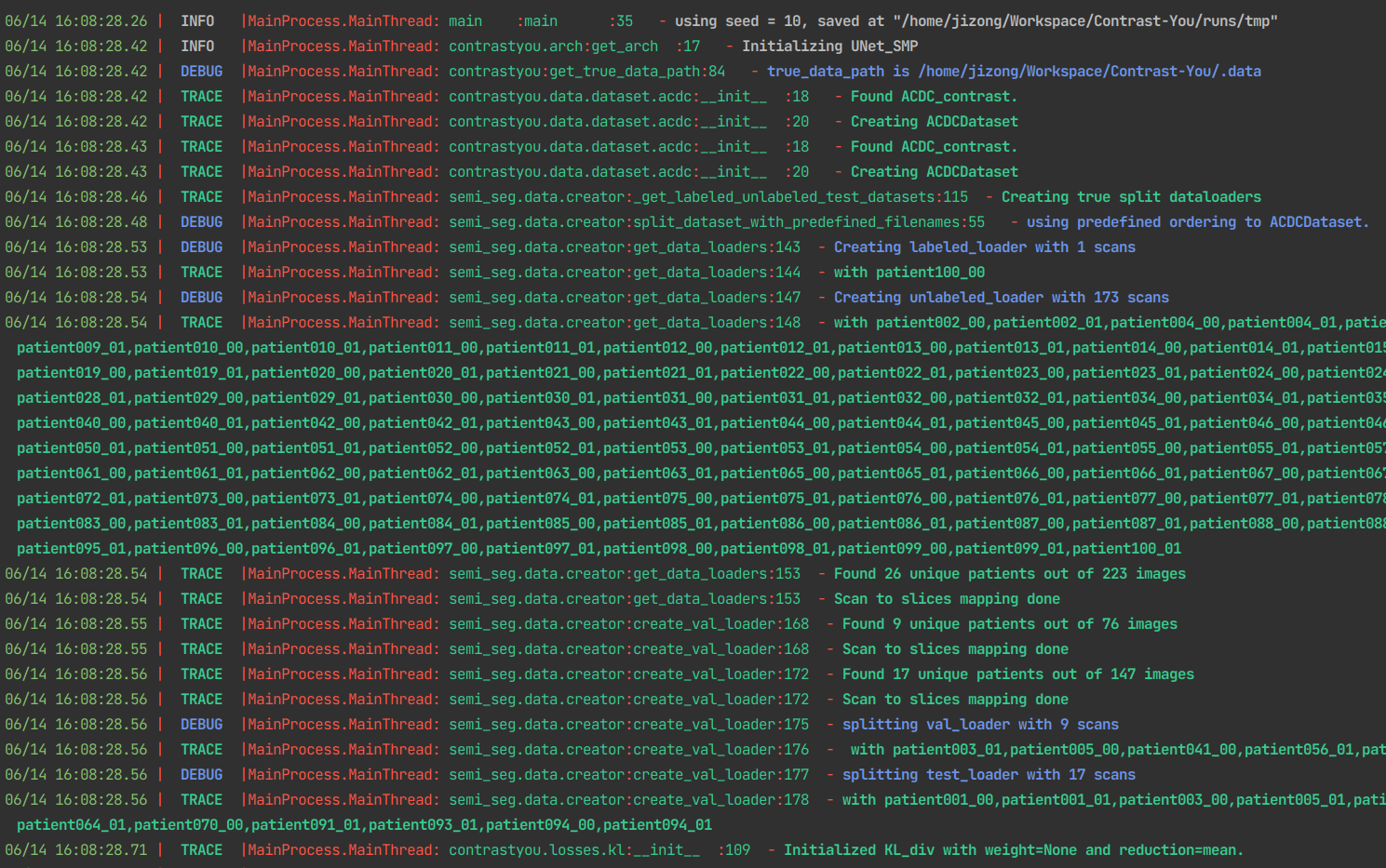 logger with trace:
logger with trace: Export LOGURU_LEVEL=TRACE
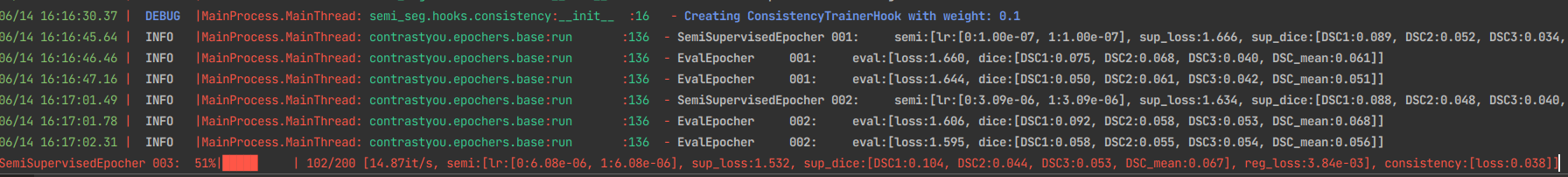
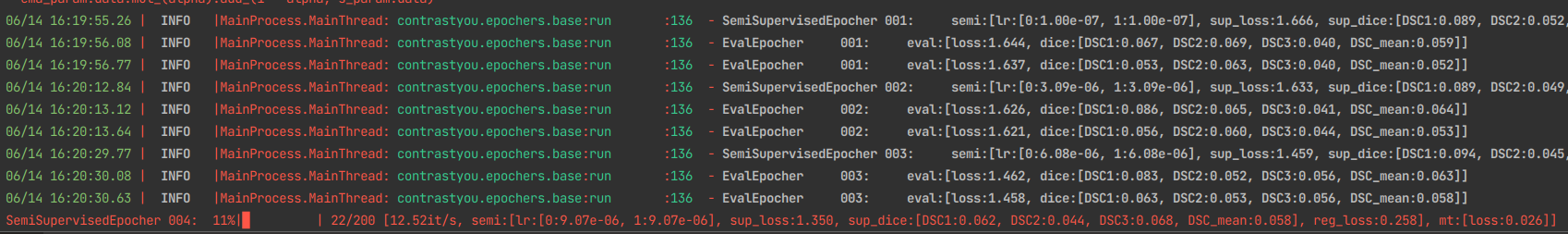
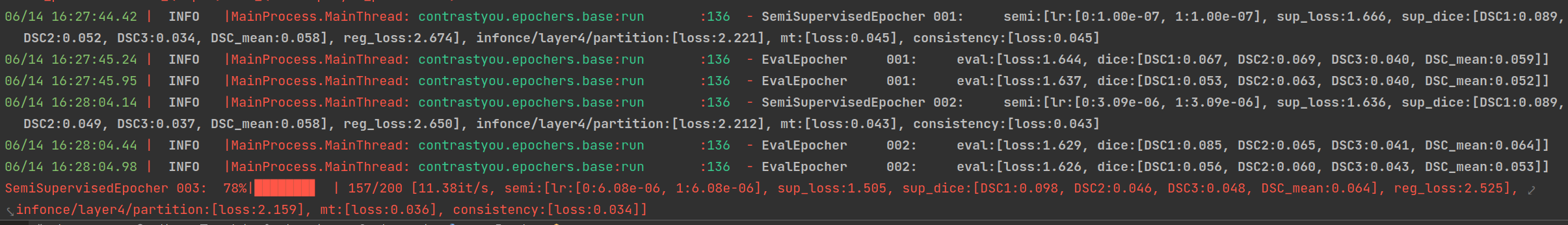
- Extendable Hooks for semi-supervised learning
with hook_registration(*hooks):
trainer.init()
if checkpoint:
trainer.resume_from_path(checkpoint)
trainer.start_training()
return trainer.inference(checkpoint_path=checkpoint)python main_nd.py -o Trainer.name=semi -p config/base.yaml config/hooks/consistency.yamlpython main_nd.py -o Trainer.name=semi -p config/base.yaml config/hooks/mt.yamlpython main_nd.py -o Trainer.name=semi -p config/base.yaml config/hooks/mt.yaml \
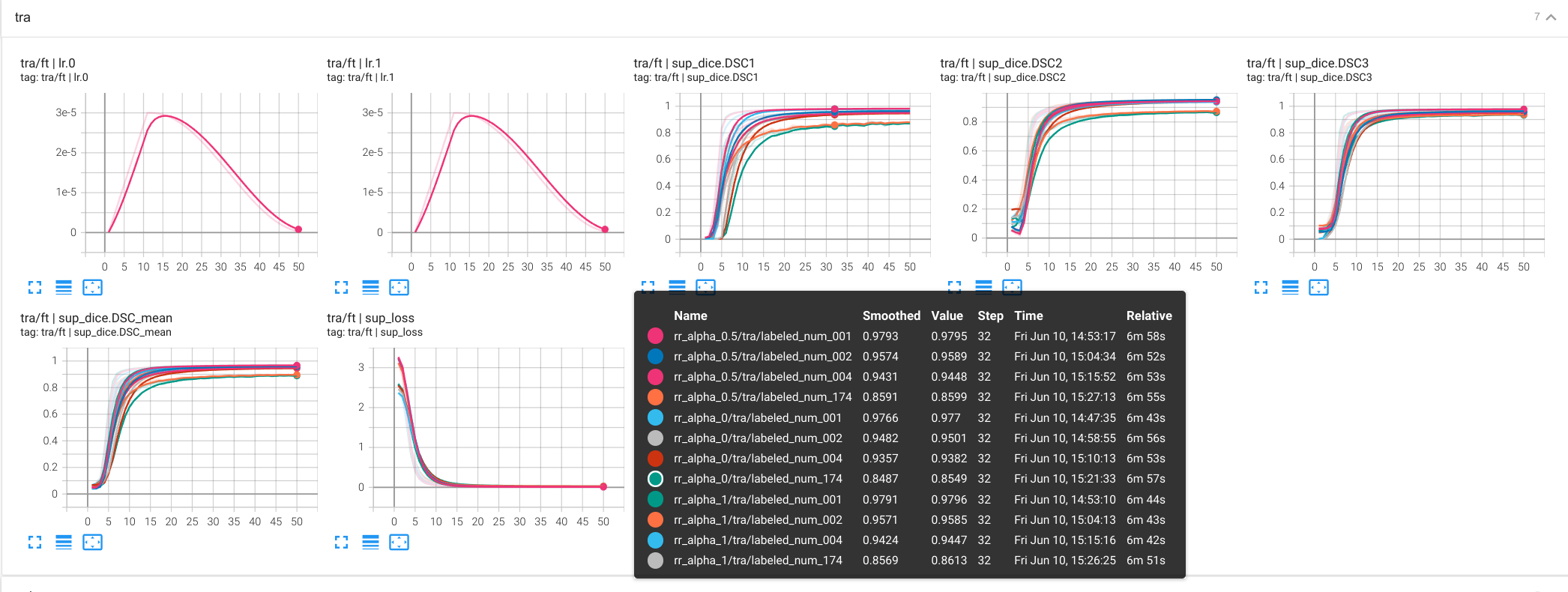
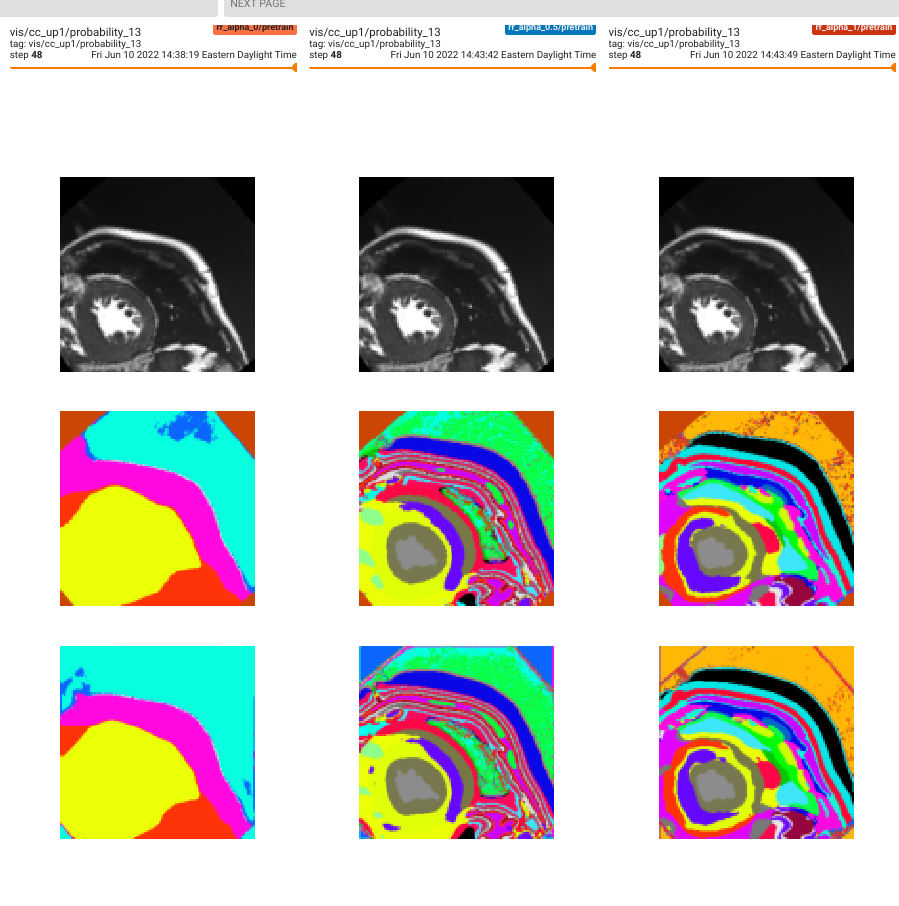
config/hooks/consistency.yaml config/hooks/infonce_encoder.yaml -o InfonceParams.feature_names=layer4 - Tenosrboard integration for experiments
@article{peng2022boundary,
title={Boundary-aware Information Maximization for Self-supervised Medical Image Segmentation},
author={Peng, Jizong and Wang, Ping and Pedersoli, Marco and Desrosiers, Christian},
journal={arXiv preprint arXiv:2202.02371},
year={2022}
}
@article{peng2021diversified,
title={Diversified Multi-prototype Representation for Semi-supervised Segmentation},
author={Peng, Jizong and Desrosiers, Christian and Pedersoli, Marco},
journal={arXiv preprint arXiv:2111.08651},
year={2021}
}
@article{peng2021self,
title={Self-paced contrastive learning for semi-supervised medical image segmentation with meta-labels},
author={Peng, Jizong and Wang, Ping and Desrosiers, Christian and Pedersoli, Marco},
journal={Advances in Neural Information Processing Systems},
volume={34},
year={2021}
}
@article{peng2021boosting,
title={Boosting semi-supervised image segmentation with global and local mutual information regularization},
author={Peng, Jizong and Pedersoli, Marco and Desrosiers, Christian},
journal={arXiv preprint arXiv:2103.04813},
year={2021}
}