Jiancheng Yang, Rui Shi, Bingbing Ni
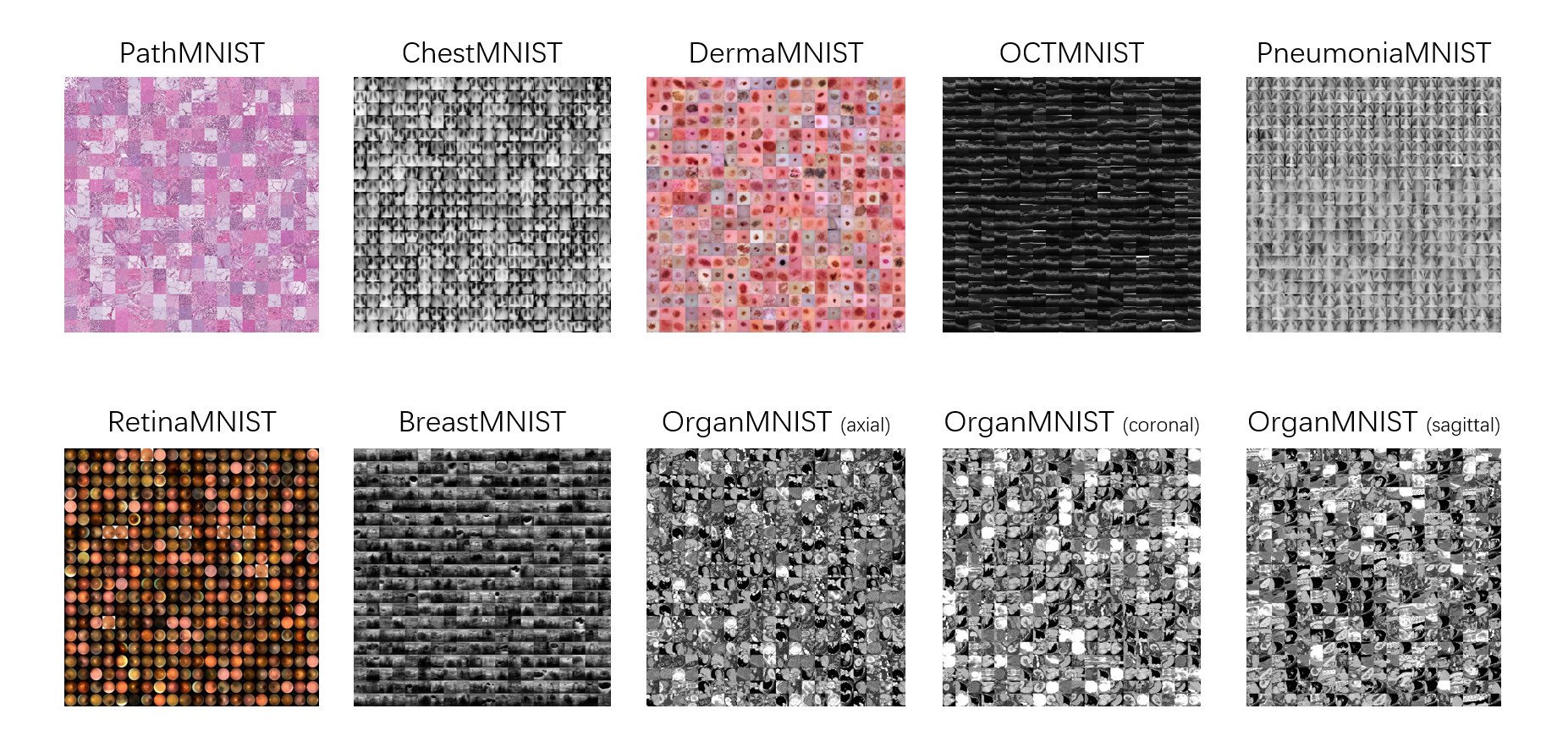
We present MedMNIST, a collection of 10 pre-processed medical open datasets. MedMNIST is standardized to perform classification tasks on lightweight 28 × 28 images, which requires no background knowledge. Covering the primary data modalities in medical image analysis, it is diverse on data scale (from 100 to 100,000) and tasks (binary/multi-class, ordinal regression and multi-label). MedMNIST could be used for educational purpose, rapid prototyping, multi-modal machine learning or AutoML in medical image analysis. Moreover, MedMNIST Classification Decathlon is designed to benchmark AutoML algorithms on all 10 datasets.
For more details, please refer to our paper:
MedMNIST Classification Decathlon: A Lightweight AutoML Benchmark for Medical Image Analysis (ISBI'21)
- Educational: Our multi-modal data, from multiple open medical image datasets with Creative Commons (CC) Licenses, is easy to use for educational purpose.
- Standardized: Data is pre-processed into same format, which requires no background knowledge for users.
- Diverse: The multi-modal datasets covers diverse data scales (from 100 to 100,000) and tasks (binary/multiclass, ordinal regression and multi-label).
- Lightweight: The small size of 28 × 28 is friendly for rapid prototyping and experimenting multi-modal machine learning and AutoML algorithms.
Please note that this dataset is NOT intended for clinical use.
medmnist/:dataset.py: PyTorch datasets and dataloaders of MedMNIST.evaluator.py: Standardized evaluation functions.info.py: Dataset informationdictfor each subset of MedMNIST.
examples/:getting_started.ipynb: Explore the MedMNIST dataset with jupyter notebook. It is ONLY intended for a quick exploration, i.e., it does not provide full training and evaluation functionalities. Please refer to our another repositoryMedMNIST/experimentsfor all experiments, including PyTorch, auto-sklearn, AutoKeras and Google AutoML Vision, together with their weights!- #TODO
getting_started_without_PyTorch.ipynb: This notebook provides snippets about how to use MedMNIST data (the.npzfiles) without PyTorch.
setup.py: The script to install medmnist as a module
Setup the required environments and install medmnist as a standard Python package:
pip install --upgrade git+https://github.com/MedMNIST/MedMNIST.git
Check whether you have isnstalled the latest version:
>>> import medmnist
>>> print(medmnist.__version__)
The code requires only common Python environments for machine learning. Basicially, it was tested with
- Python 3 (Anaconda 3.6.3 specifically)
- PyTorch==0.3.1
- numpy==1.18.5, pandas==0.25.3, scikit-learn==0.22.2, Pillow==8.0.1, fire
Higher (or lower) versions should also work (perhaps with minor modifications).
It is suggested to use our PyTorch dataset code to parse the .npz files; However, you are free to parse them with your own code (without PyTorch or even without Python!), as they are only standard NumPy serialization files. Please refer to getting_started_without_PyTorch.ipynb, which provides snippets about how to use MedMNIST data (the .npz files) without PyTorch.
Please download the dataset(s) via Zenodo. You could also use our code to download automatically.
The MedMNIST dataset contains several subsets. Each subset (e.g., pathmnist.npz) is comprised of 6 keys: train_images, train_labels, val_images, val_labels, test_images and test_labels.
train_images/val_images/test_images:Nx 28 x 28 x 3 for RGB,Nx 28 x 28 for gray-scale.Ndenotes the number of samples.train_labels/val_labels/test_labels:NxL.Ndenotes the number of samples.Ldenotes the number of task labels; for single-label (binary/multi-class) classification,L=1, and{0,1,2,3,..,C}denotes the category labels (C=1for binary); for multi-label classificationL!=1, e.g.,L=14forchestmnist.npz.
-
List all available datasets:
python -m medmnist available -
Download all available datasets:
python -m medmnist download -
Delete all downloaded npz from root:
python -m medmnist clean -
Print the dataset details given a subset flag:
python -m medmnist info --flag=xxxmnist -
Save the dataset as standard figures, which could be used for AutoML tools, e.g., Google AutoML Vision:
python -m medmnist save --flag=xxxmnist --folder=tmp/ -
Download the dataset manually or automatically (by setting
download=Trueindataset.py). -
Explore the MedMNIST dataset with jupyter notebook (
getting_started.ipynb), and train basic neural networks in PyTorch. -
If you do not use PyTorch, go to
getting_started_without_PyTorch.ipynb, which provides snippets about how to use MedMNIST data (the.npzfiles) without PyTorch. -
Please refer to our another repository
MedMNIST/experimentsfor all experiments, including PyTorch, auto-sklearn, AutoKeras and Google AutoML Vision together with their weights!
If you find this project useful, please cite our paper as:
Jiancheng Yang, Rui Shi, Bingbing Ni. "MedMNIST Classification Decathlon: A Lightweight AutoML Benchmark for Medical Image Analysis," IEEE 18th International Symposium on Biomedical Imaging (ISBI), 2021.
or using the bibtex:
@inproceedings{medmnistv1,
title={MedMNIST Classification Decathlon: A Lightweight AutoML Benchmark for Medical Image Analysis},
author={Yang, Jiancheng and Shi, Rui and Ni, Bingbing},
booktitle={IEEE 18th International Symposium on Biomedical Imaging (ISBI)},
pages={191--195},
year={2021}
}
Please also cite the corresponding paper of source data if you use any subset of MedMNIST as per the description in the project page.
The code is under Apache-2.0 License.
The datasets are under Creative Commons (CC) Licenses in general. Each subset uses the same license as that of the source dataset, please refer to the project page for details.