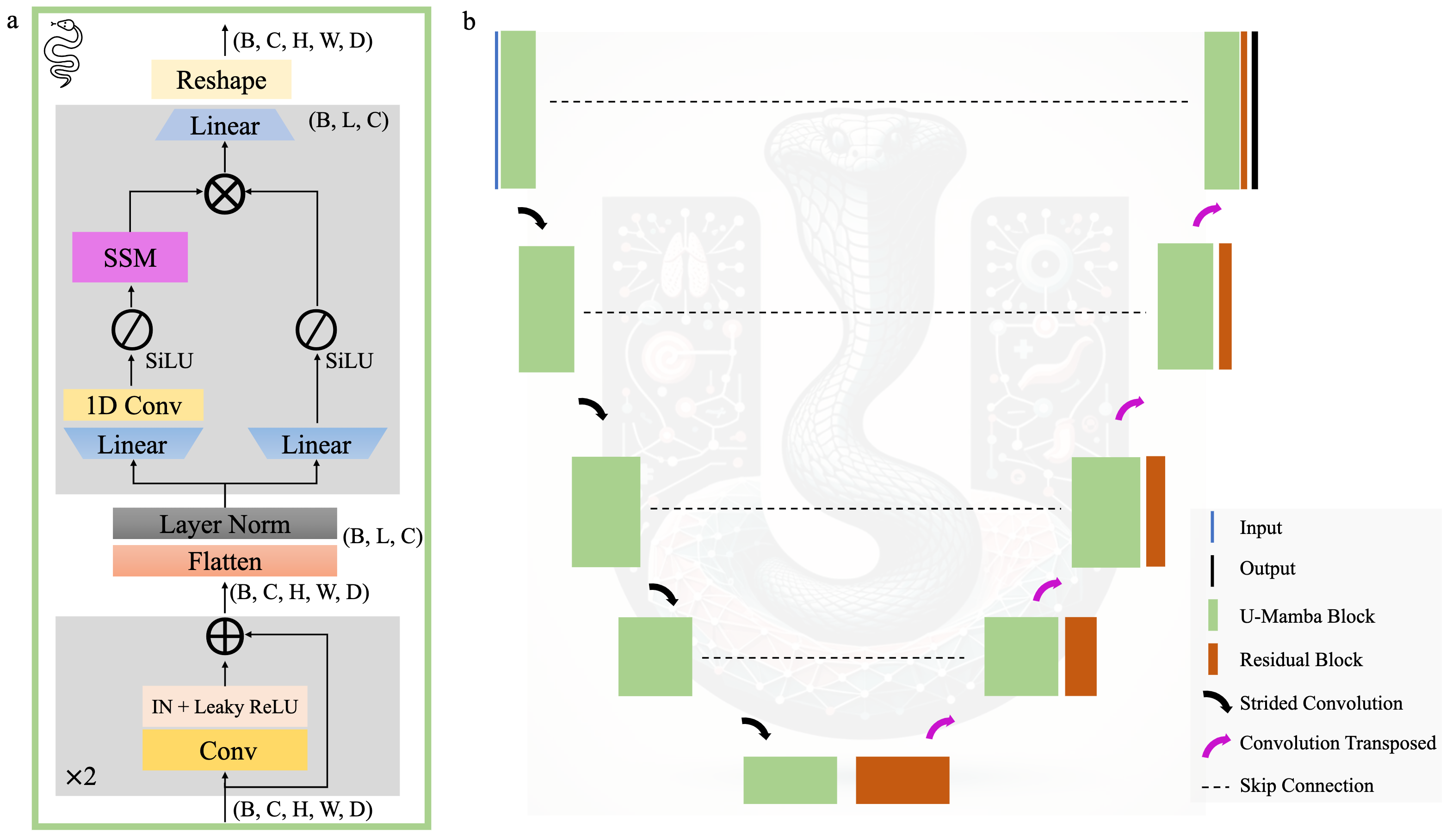
Official repository for U-Mamba: Enhancing Long-range Dependency for Biomedical Image Segmentation. Welcome to join our mailing list to get updates.
Requirements: Ubuntu 20.04, CUDA 11.8
- Create a virtual environment:
conda create -n umamba python=3.10 -yandconda activate umamba - Install Pytorch 2.0.1:
pip install torch==2.0.1 torchvision==0.15.2 --index-url https://download.pytorch.org/whl/cu118 - Install Mamba:
pip install causal-conv1d>=1.2.0andpip install mamba-ssm --no-cache-dir - Download code:
git clone https://github.com/bowang-lab/U-Mamba cd U-Mamba/umambaand runpip install -e .
sanity test: Enter python command-line interface and run
import torch
import mamba_ssmvisual_seg.mp4
Download dataset here and put them into the data folder. U-Mamaba is built on the popular nnU-Net framework. If you want to train U-Mamba on your own dataset, please follow this guideline to prepare the dataset.
nnUNetv2_plan_and_preprocess -d DATASET_ID --verify_dataset_integrity- Train 2D
U-Mamba_Botmodel
nnUNetv2_train DATASET_ID 2d all -tr nnUNetTrainerUMambaBot- Train 2D
U-Mamba_Encmodel
nnUNetv2_train DATASET_ID 2d all -tr nnUNetTrainerUMambaEnc- Train 3D
U-Mamba_Botmodel
nnUNetv2_train DATASET_ID 3d_fullres all -tr nnUNetTrainerUMambaBot- Train 3D
U-Mamba_Encmodel
nnUNetv2_train DATASET_ID 3d_fullres all -tr nnUNetTrainerUMambaEnc- Predict testing cases with
U-Mamba_Botmodel
nnUNetv2_predict -i INPUT_FOLDER -o OUTPUT_FOLDER -d DATASET_ID -c CONFIGURATION -f all -tr nnUNetTrainerUMambaBot --disable_tta- Predict testing cases with
U-Mamba_Encmodel
nnUNetv2_predict -i INPUT_FOLDER -o OUTPUT_FOLDER -d DATASET_ID -c CONFIGURATION -f all -tr nnUNetTrainerUMambaEnc --disable_tta
CONFIGURATIONcan be2dand3d_fullresfor 2D and 3D models, respectively.
- Path settings
The default data directory for U-Mamba is preset to U-Mamba/data. Users with existing nnUNet setups who wish to use alternative directories for nnUNet_raw, nnUNet_preprocessed, and nnUNet_results can easily adjust these paths in umamba/nnunetv2/path.py to update your specific nnUNet data directory locations, as demonstrated below:
# An example to set other data path,
base = '/home/user_name/Documents/U-Mamba/data'
nnUNet_raw = join(base, 'nnUNet_raw') # or change to os.environ.get('nnUNet_raw')
nnUNet_preprocessed = join(base, 'nnUNet_preprocessed') # or change to os.environ.get('nnUNet_preprocessed')
nnUNet_results = join(base, 'nnUNet_results') # or change to os.environ.get('nnUNet_results')- AMP could lead to nan in the Mamba module. We also provide a trainer without AMP: https://github.com/bowang-lab/U-Mamba/blob/main/umamba/nnunetv2/training/nnUNetTrainer/nnUNetTrainerUMambaEncNoAMP.py
@article{U-Mamba,
title={U-Mamba: Enhancing Long-range Dependency for Biomedical Image Segmentation},
author={Ma, Jun and Li, Feifei and Wang, Bo},
journal={arXiv preprint arXiv:2401.04722},
year={2024}
}
We acknowledge all the authors of the employed public datasets, allowing the community to use these valuable resources for research purposes. We also thank the authors of nnU-Net and Mamba for making their valuable code publicly available.