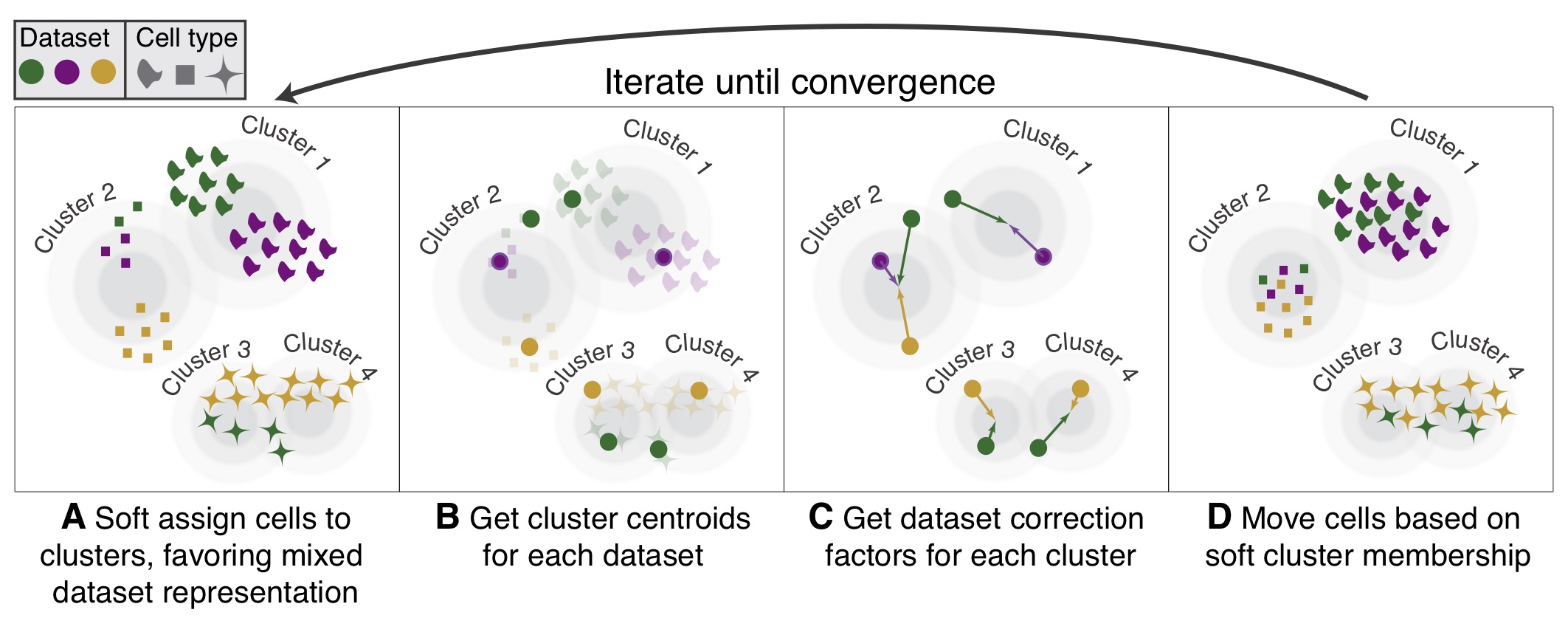
Scalable integration of single cell RNAseq data for batch correction and meta analysis
Check out the latest preprint of Harmony on bioRxiv
Harmony has been tested on R versions >= 3.4. Please consult the DESCRIPTION file for more details on required R packages. Harmony has been tested on Linux, OS X, and Windows platforms.
To run Harmony, open R and install directly from github using the following commands:
library(devtools)
install_github("immunogenomics/harmony")
Installation may include compiling C++ code from source, so it can take a few minutes.
We made it easy to run Harmony in most common R analysis pipelines.
Check out this vignette for a quick start tutorial.
The Harmony algorithm iteratively corrects PCA embeddings. To input your own low dimensional embeddings directly, set do_pca=FALSE. Harmony is packaged with a small dataset
library(harmony)
my_harmony_embeddings <- HarmonyMatrix(my_pca_embeddings, meta_data, "dataset", do_pca=FALSE)
You can also run Harmony on a sparse matrix of library size normalized expression counts. Harmony will scale these counts, run PCA, and finally perform integration.
library(harmony)
my_harmony_embeddings <- HarmonyMatrix(normalized_counts, meta_data, "dataset")
You can run Harmony within your Seurat workflow. You'll only need to make two changes to your code.
- Run Harmony with the
RunHarmony()function - In downstream analyses, use the Harmony embeddings instead of PCA.
For example, run Harmony and then UMAP in two lines.
seuratObj <- RunHarmony(seuratObj, "dataset")
seuratObj <- RunUMAP(seuratObj, reduction = "harmony")
For details, check out these vignettes:
You can run Harmony with functions from the MUDAN package. For more, details, check out this vignette.
Harmony can integrate over multiple covariates. To do this, specify a vector covariates to integrate.
my_harmony_embeddings <- HarmonyMatrix(my_pca_embeddings, meta_data, c("dataset", "donor", "batch_id"), do_pca=FALSE)
Do the same with your Seurat object:
seuratObject <- RunHarmony(seuratObject, c("dataset", "donor", "batch_id"))
The examples above all return integrated PCA embeddings. We created a more advanced tutorial that explores the internal data structures used in the Harmony algorithm.
Code to reproduce Harmony results from the Korsunsky et al 2019 manuscript will be made available on github.com/immunogenomics/harmony2019.