Biomedical imaging is vital for the diagnosis and treatment of various medical conditions, yet the effective integration of deep learning technologies into this field presents challenges. Traditional methods often struggle to efficiently capture the spatial characteristics and intricate structural features of 3D volumetric medical images, limiting memory utilization and model adaptability. To address this, we introduce a Linked Memory Token Turing Machine (LMTTM), which utilizes external linked memory to efficiently process spatial dependencies and structural complexities within 3D volumetric medical images, aiding in accurate diagnoses. LMTTM can efficiently record the features of 3D volumetric medical images in an external linked memory module, enhancing complex image classification through improved feature accumulation and reasoning capabilities. Our experiments on six 3D volumetric medical image datasets from the MedMNIST v2 demonstrate that our proposed LMTTM model achieves average ACC of 82.4%, attaining state-of-the-art (SOTA) performance. Moreover, ablation studies confirmed that the Linked Memory outperforms its predecessor, TTM's original Memory, by up to 5.7%, highlighting LMTTM's effectiveness in 3D volumetric medical image classification and its potential to assist healthcare professionals in diagnosis and treatment planning.
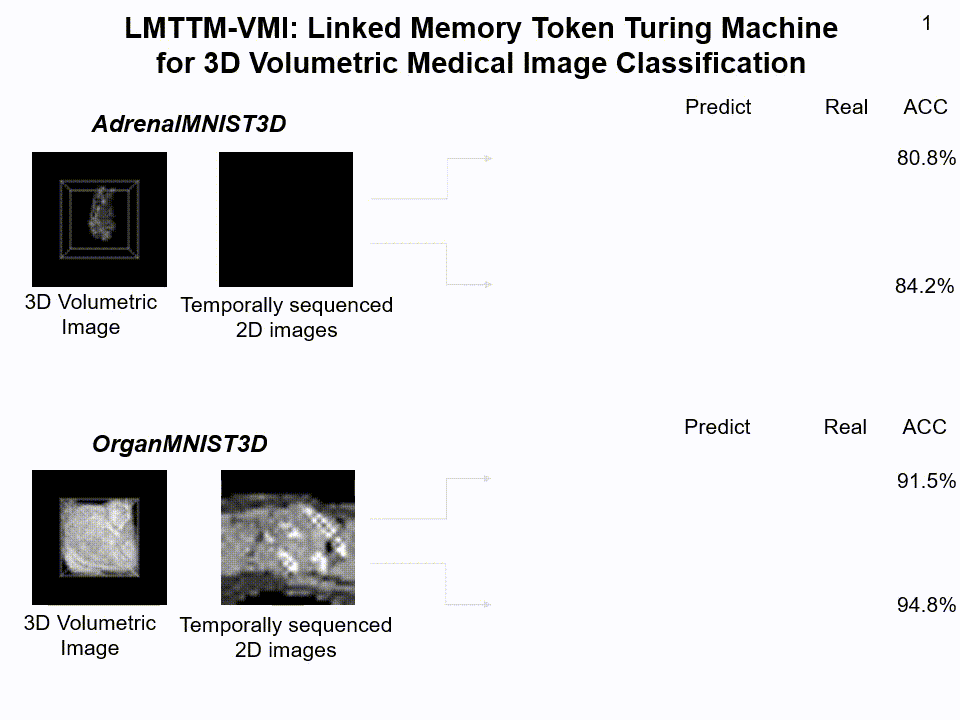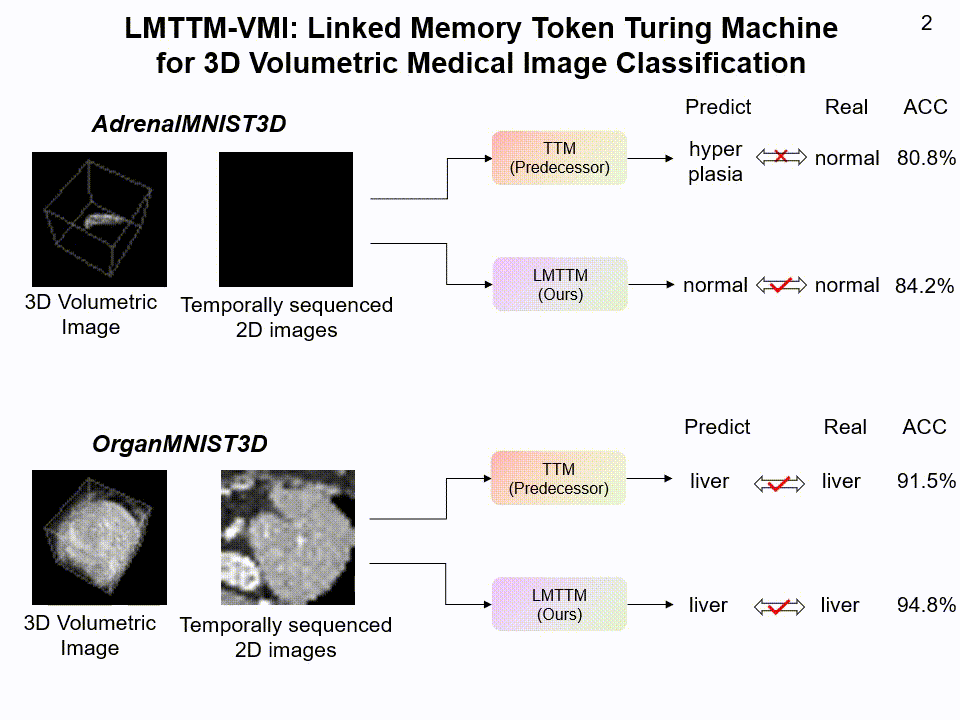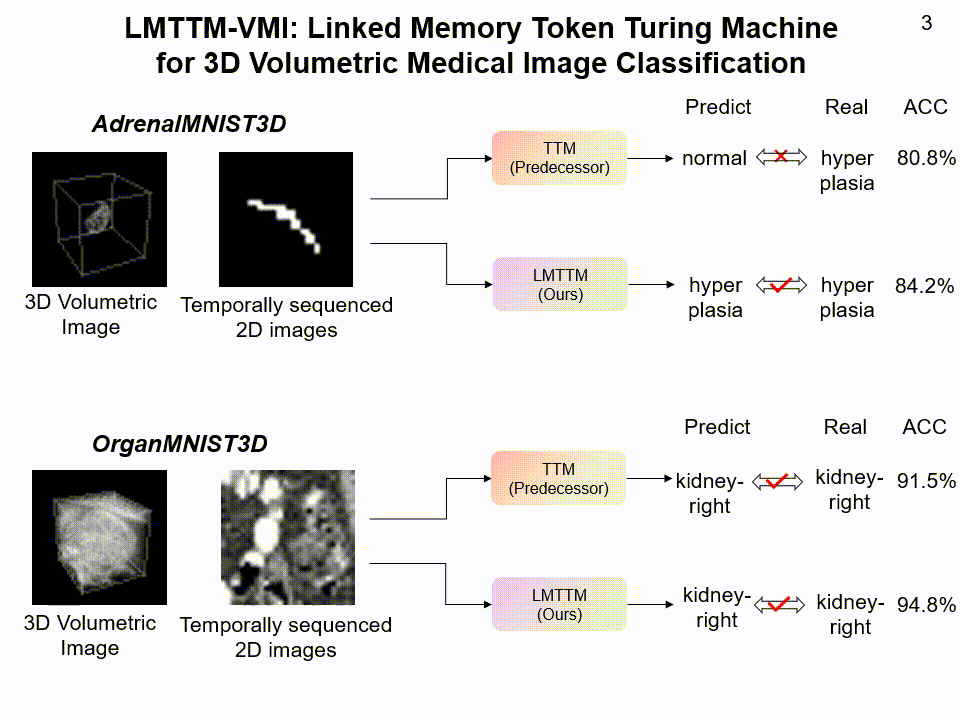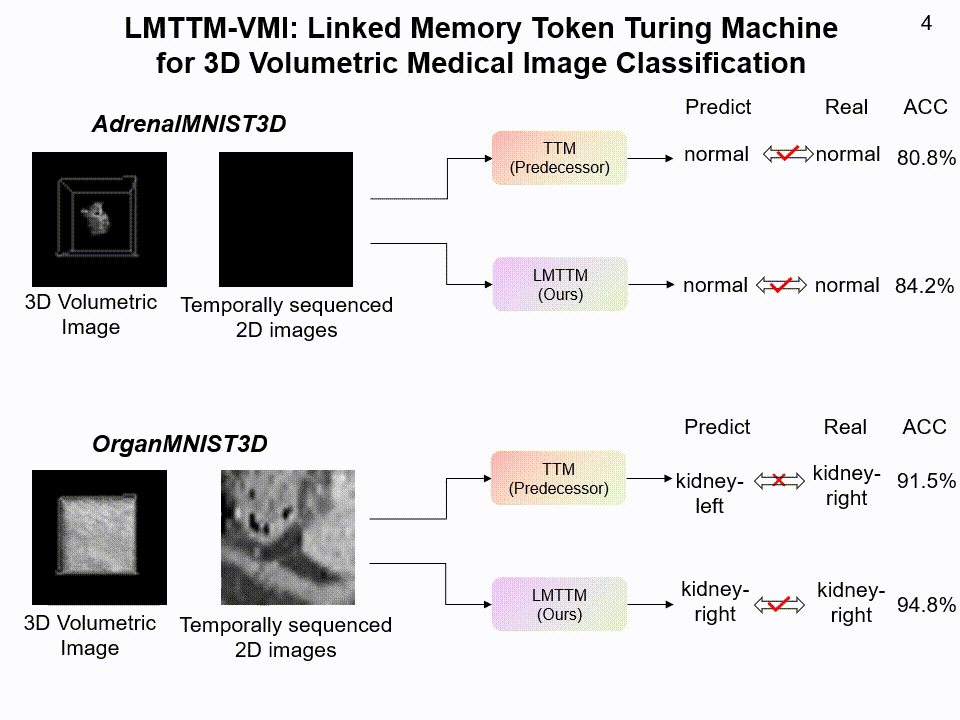
LMTTM-VMI on AdrenalMNIST3D and OrganMNIST3D dataset
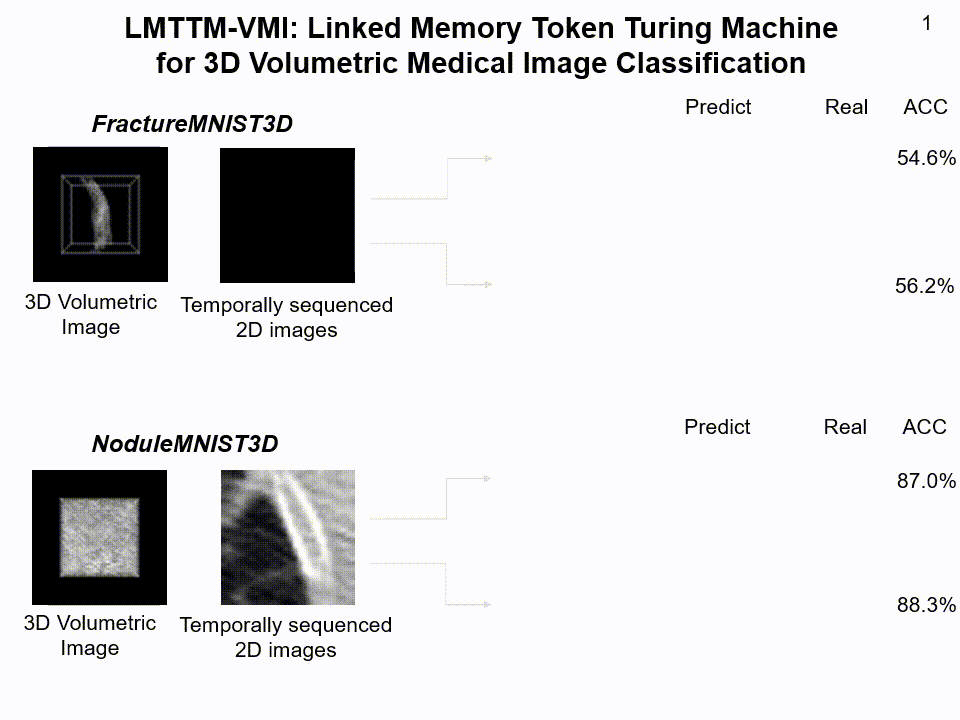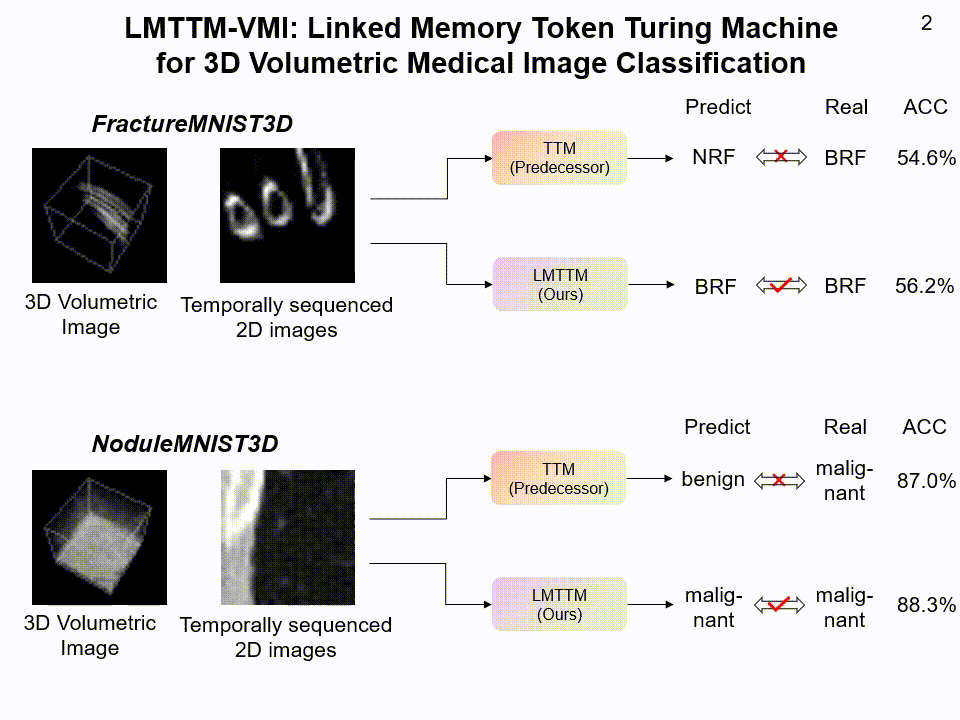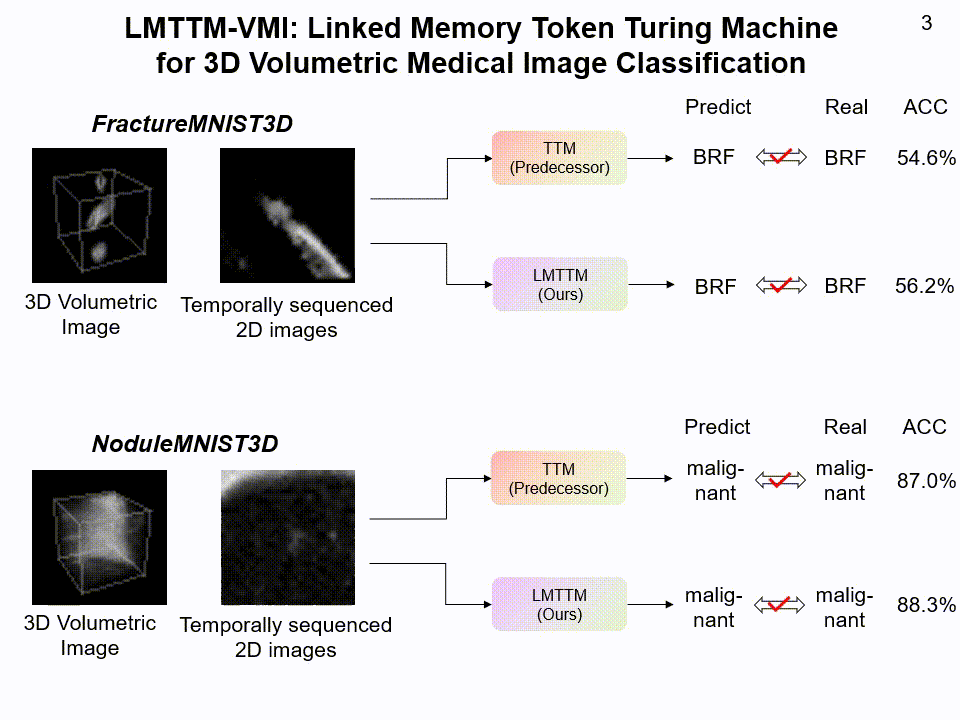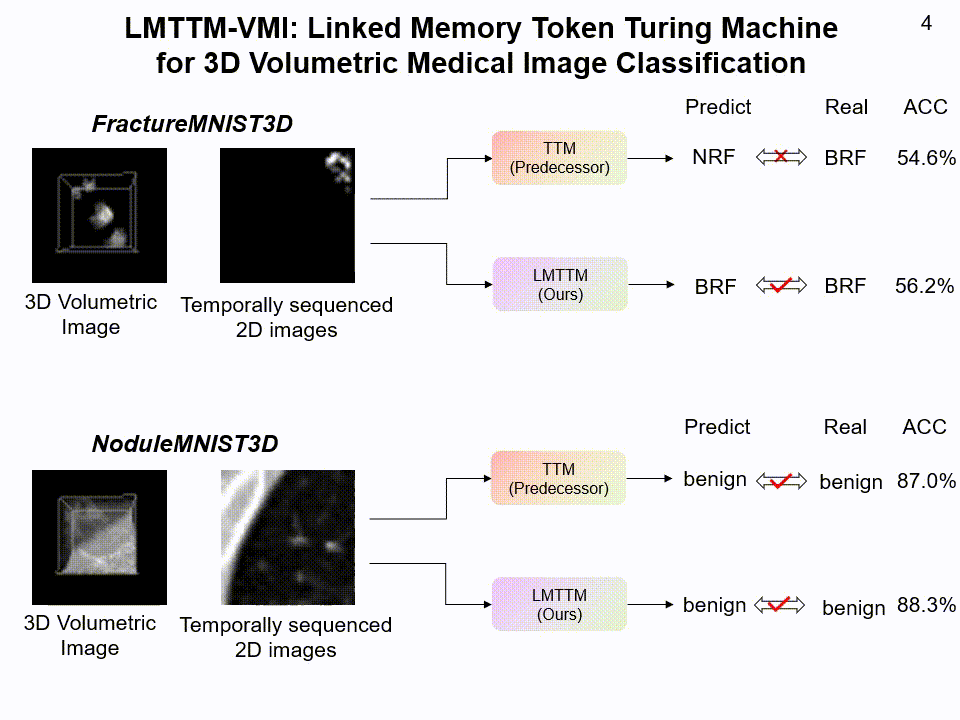
LMTTM-VMI on FractureMNIST3D and NoduleMNIST3D dataset
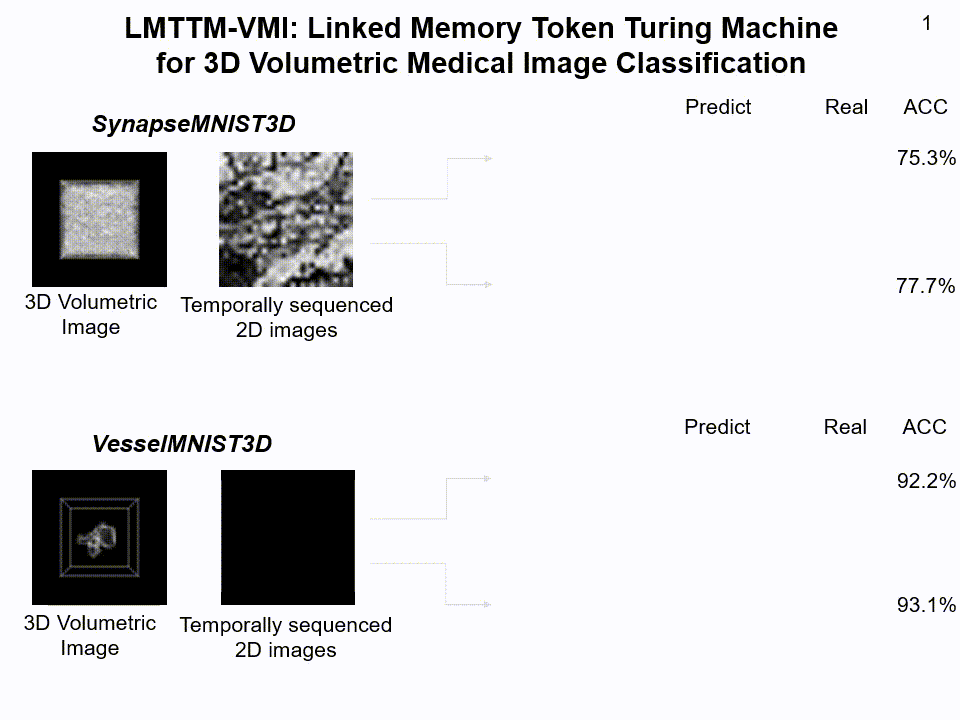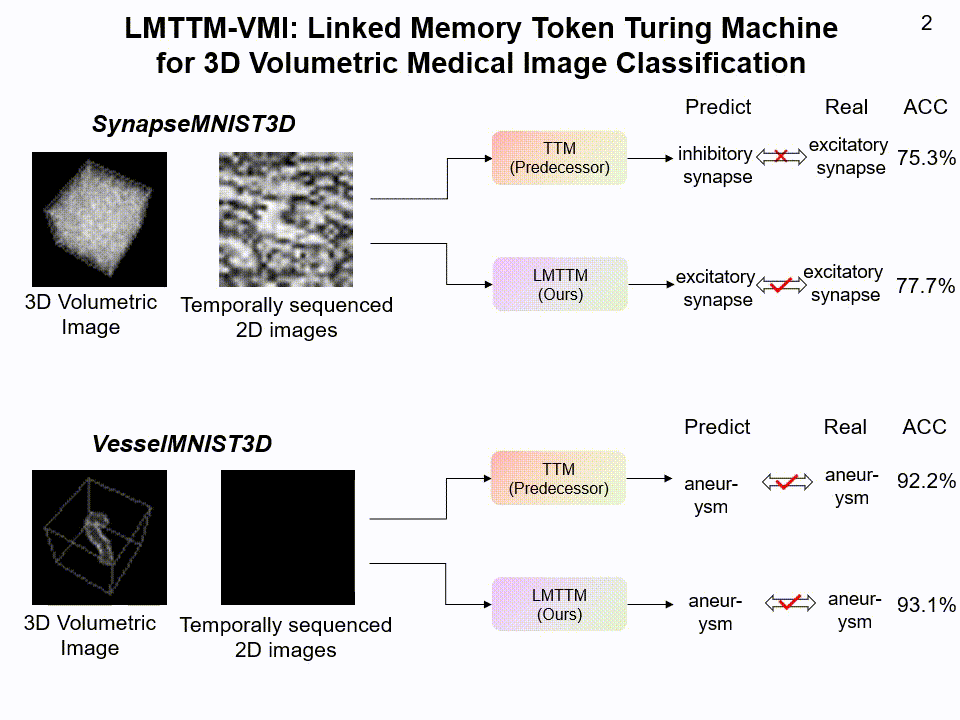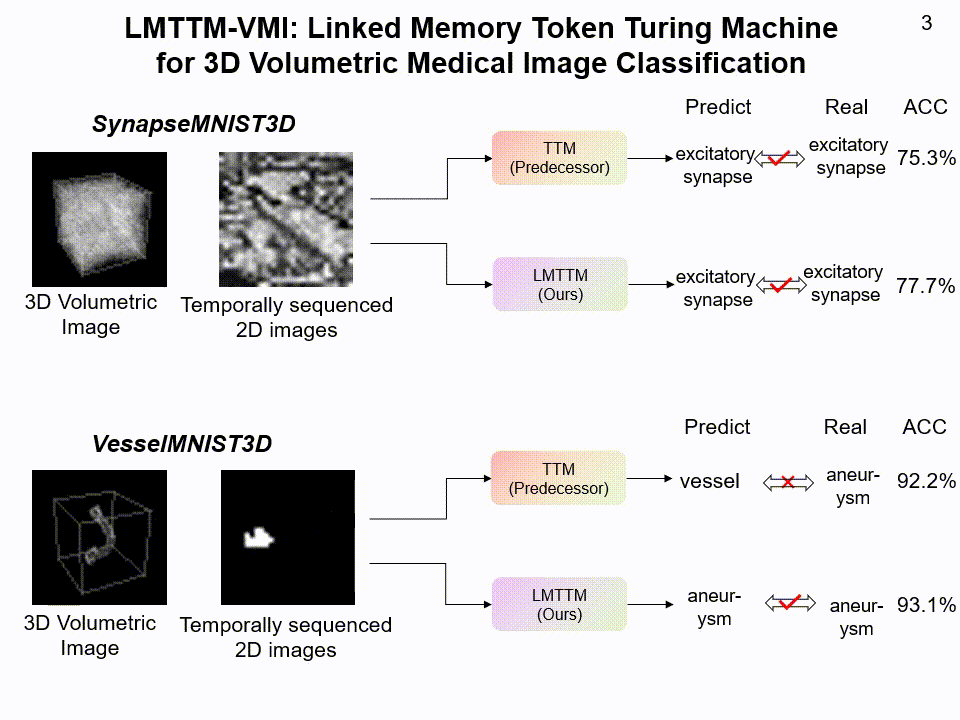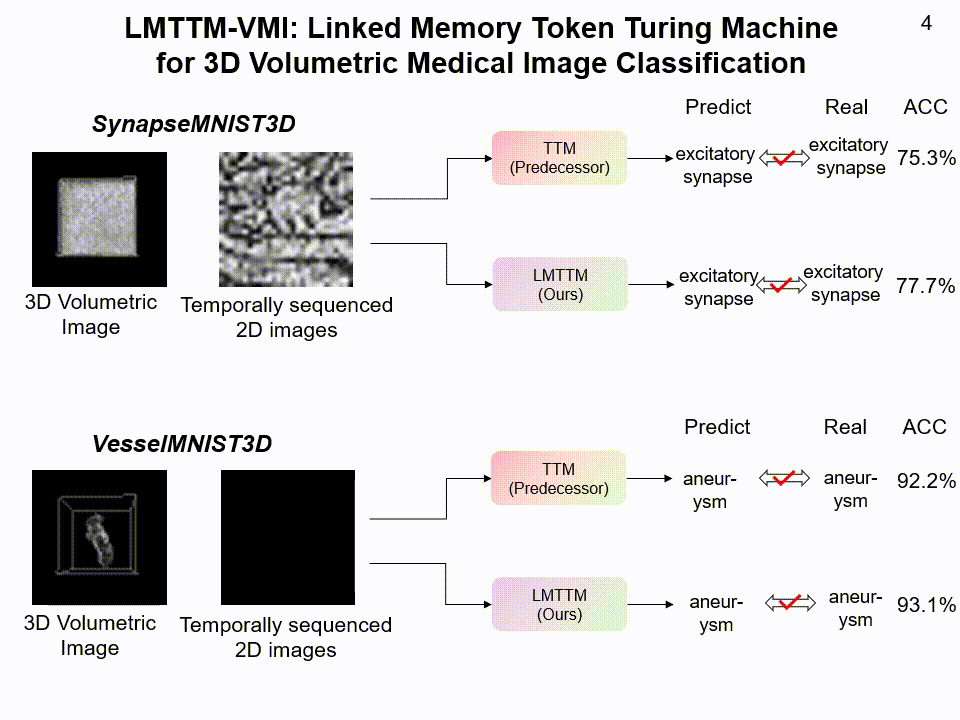
LMTTM-VMI on SynapseMNIST3D and VesselMNIST3D dataset
represents memory blocks at different times, the three moments of memory are the past moment, this moment, and the next moment.
represents the input at moment t.
represents the output features obtained after MDU output. It is used for result classification and writing to memory.
represents the size of the memory block, respectively the memory block length and dimension.
denotes the feature information read from memory at different moments in time.
Schematics of the Linked Memory Token Turing Machine (LMTTM). On the left, the network's processing at time stamp and its adjacent blocks
and
are synergistically processed by the Tri-Temporal Memory Collaborative (T-TMC) module, which we will refer to as Simplified Memory. Simplified memory, along with preprocessed input tokens, is read through the Read module, and the extracted information is then refined into more efficient output tokens by the Memory Distillation Unit(MDU) for image classification. The simplified Memory, input tokens, and output tokens are then written in the subsequent Memory block
via the Write module, completing the LMTTM cycle for time stamp
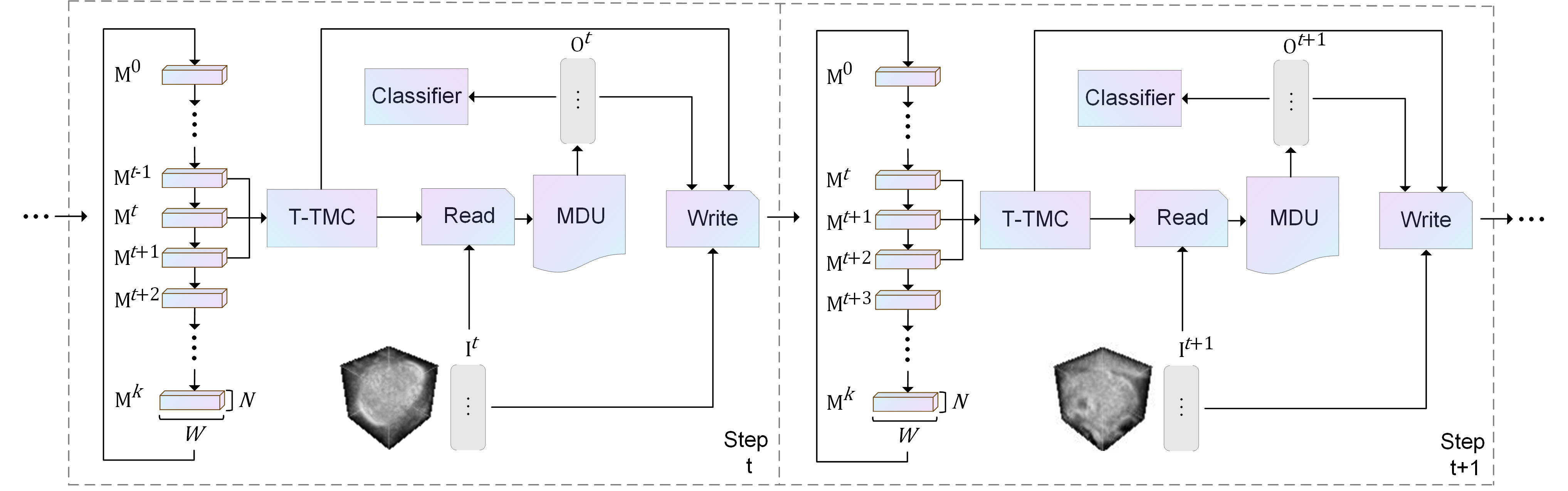
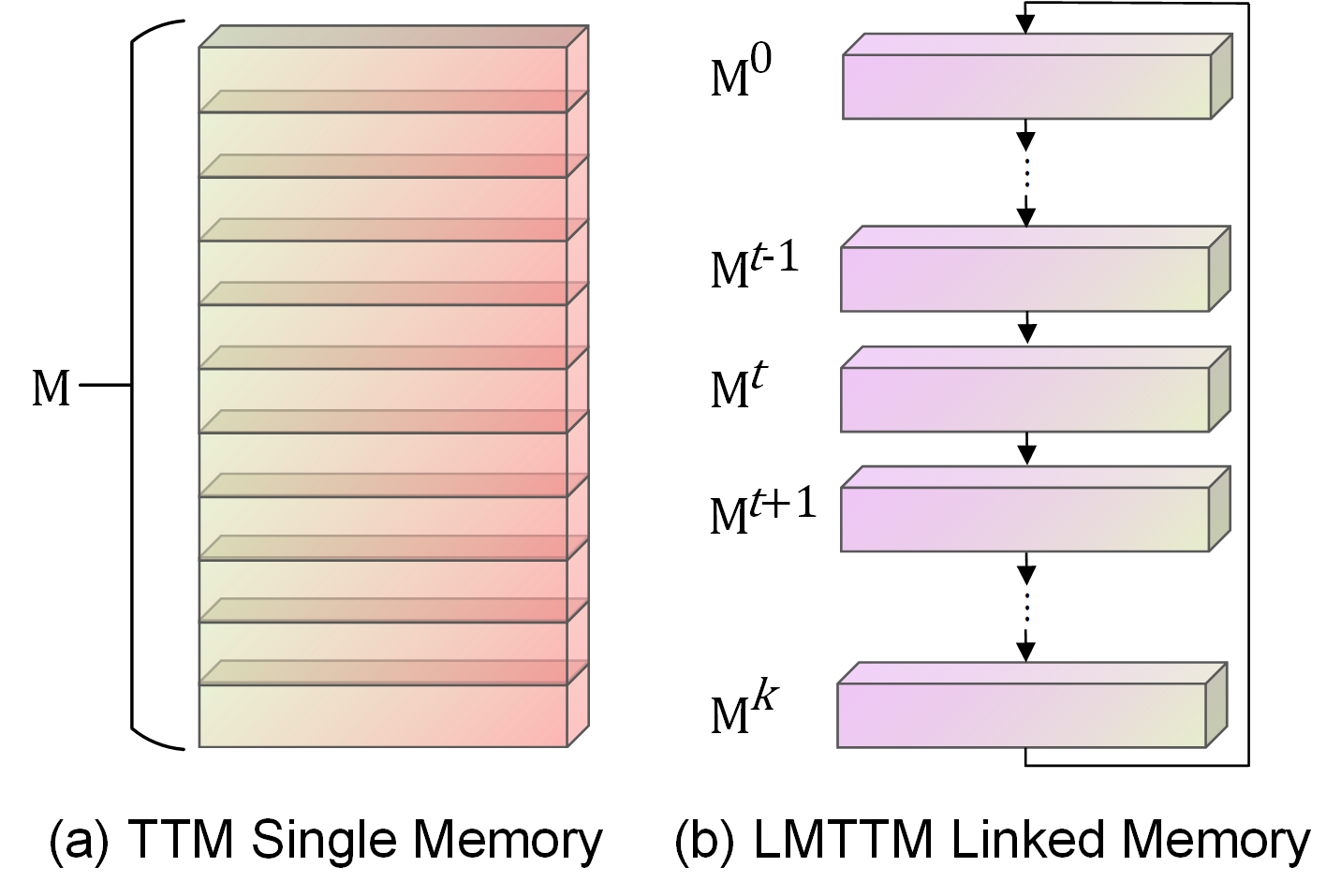
Illustration of different memory. (a) is the memory structure of TTM, where the entire memory block is iteratively updated during the interaction. (b) is the memory structure of our LMTTM, which can be viewed as a chain of k memory units linked together, and is iteratively updated one memory unit at a time during the interaction.
Illustration of different Reads. (a) is the entire block of memory that will be read by the TTM. (b) is the coordinated read operation for the current moment 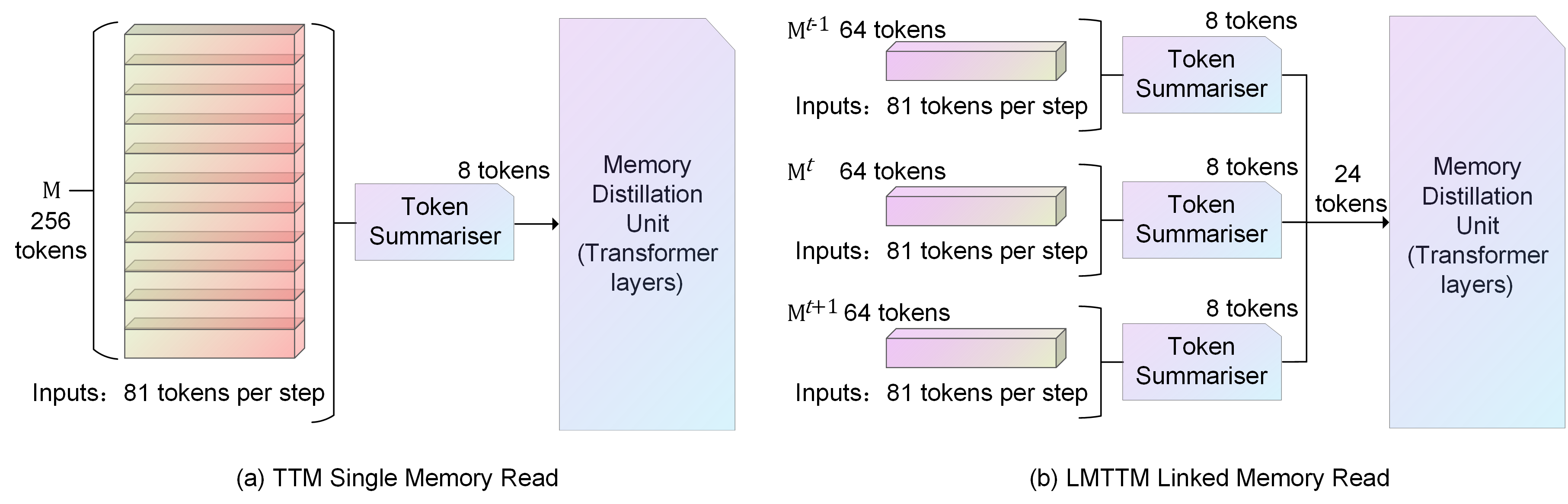
In the LMTTM architecture, the processing unit is designed as , which receives the tokens
and processes them. The MDU outputs a set of vectors
containing the
to enhance the classification capability of the model, which is implemented as
, where
is the weight matrix of the fully connected layer used to map
to the input space of the classifier. This produces the final classification prediction.
Illustration of different Writes. (a) is the entire block of memories from the current moment in TTM being written to the next moment in the entire block. (b) is the entire block of tri-temporal memories from the current moment 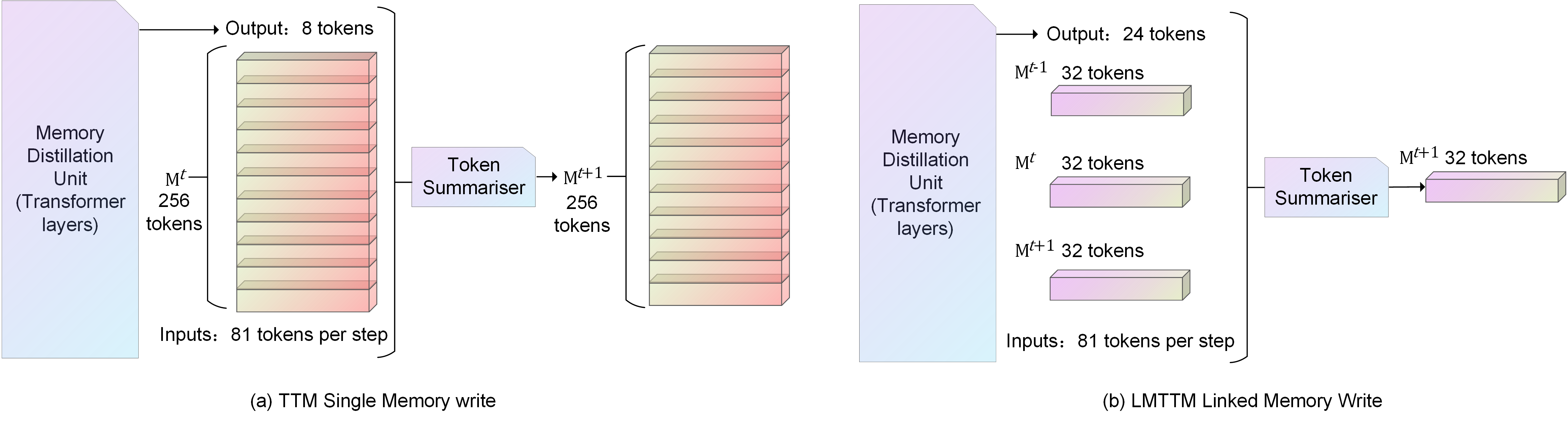
| Name | Version |
|---|---|
| Python | 3.8 |
| CUDA | >=10.1 |
| Pytorch | 1.12.1 |
The rest of the environment is installed with the following command
cd <project path>
pip install -r requirement.txtClone the repository
git clone <repository url>MedMNIST v2 contains a collection of six preprocessed 3D volumetric medical image datasets. It is designed to be educational, standardized, diverse, and lightweight and can be used as a general classification benchmark in 3D volumetric medical image analysis.
The program will automatically download the corresponding dataset at runtime.
For different datasets and tasks, different parameters need to be configured.
The parameters are configured in <path>\config\base.json.
To run the program, you need to change the dataset_name field in base.json to the name of the corresponding dataset, which can be OrganMNIST3D, SynapseMNIST3D, AdrenalMNIST3D, FractureMNIST3D, NoduleMNIST3D, VesselMNIST3D, and also in out_class_num you need to change the number of classes in the corresponding dataset. VesselMNIST3D, also in out_class_num you need to modify the number of classes in the corresponding dataset.
Modify the dataset_name, out_class_num and epoch parameters in the configuration file <path>\config\base.json.
And then activate your virtual environment, followed by executing python train.py base.json.
As with the TRAIN process, modify the corresponding parameters, then activate the virtual environment and execute python predict.py base.json
This work is based on TTM(Token Turing Machine) and inspired by CMN (Collaborative Memory Network).
LMTTM's code is released under the Apache License 2.0. A permissive license whose main conditions require preservation of copyright and license notices. Contributors provide an express grant of patent rights. Licensed works, modifications, and larger works may be distributed under different terms and without source code.See LICENSE for further details.