- Team CGIP
- Ho Yeon Lim and Min Chang Kim
- Please press the star!
- This repository contains the code for the tooth segmentation algorithm that won first place at Miccai2022 3D Teeth Scan Segmentation and Labeling Challenge.
- Official Paper for Challenge: 3DTeethSeg'22: 3D Teeth Scan Segmentation and Labeling Challenge.
- We used the dataset shared in the challenge git repository.
- If you only want the inference code, or if you want to use the same checkpoints that we used in the challenge, you can jump to challenge_branch in this repository.
- Most of the code has been modified. It may be significantly different from the code you downloaded before June 20th. If you downloaded this repository, we recommend download it again.
- There may be a lot of errors in the initial code. If you encounter any errors, please post them on the issue board, and we will promptly address and fix them(Im sorry for the slow resposes to issues because of personal reasons)...
- Please post your questions to the issue board as it's hard to see them in emails.
- We used the dataset shared in the challenge git repository. For more information about the data, check out the link.
- Dataset consists of dental mesh obj files and corresponding ground truth json files.
- You can also download the challenge training split data in google drive(our codes are based on this data).
- After you download and unzip these zip files, merge
3D_scans_per_patient_obj_files.zipand3D_scans_per_patient_obj_files_b2.zip. The parent path of these obj files isdata_obj_parent_directory. - Apply the same to the ground truth json files(
ground-truth_labels_instances.zipandground-truth_labels_instances_b2.zip. The parent path of these json files isdata_json_parent_directory). - The directory structure of your data should look like below..
--data_obj_parent_directory ----00OMSZGW ------00OMSZGW_lower.obj ------00OMSZGW_upper.obj ----0EAKT1CU ------0EAKT1CU_lower.obj ------0EAKT1CU_upper.obj and so on.. --data_json_parent_directory ----00OMSZGW ------00OMSZGW_lower.json ------00OMSZGW_upper.jsno ----0EAKT1CU ------0EAKT1CU_lower.json ------0EAKT1CU_upper.json and so on..
- After you download and unzip these zip files, merge
- If you have your dental mesh data, you can use it.
- For training, you have to execute the
preprocess_data.pyto save the farthest point sampled vertices of the mesh (.obj) files. - Here is an example of how to execute
preprocess_data.py.python preprocess.py --source_obj_data_path data_obj_parent_directory \ --source_json_data_path data_json_parent_directory \ --save_data_path path/for/preprocessed_data
- We offer six models(tsegnet | tgnet(ours) | pointnet | pointnetpp | dgcnn | pointtransformer).
- For experiment tracking, we use wandb. Please replace "entity" with your own wandb ID in the
get_default_config functionoftrain_configs/train_config_maker.py. - Due to the memory constraints, all functions have been implemented based on batch size 1 (minimum 11GB GPU RAM required). If you want to change the batch size to a different value, you will need to modify most of the functions by yourself.
- The tgnet is our 3d tooth segmentation method. Please refer to the challenge paper for an explanation of the methodology.
- You should first train the Farthest Point Sampling model and then train the Boundary Aware Point Sampling model.
- First, train the Farthest Point Sampling model as follows.
start_train.py \ --model_name "tgnet_fps" \ --config_path "train_configs/tgnet_fps.py" \ --experiment_name "your_experiment_name" \ --input_data_dir_path "path/for/preprocessed_data" \ --train_data_split_txt_path "base_name_train_fold.txt" \ --val_data_split_txt_path "base_name_val_fold.txt"- Input the preprocessed data directory path into the
--input_data_dir_path. - You can provide the train/validation split text files through
--train_data_split_txt_pathand--val_data_split_txt_path. You can either use the provided text files from the above dataset drive link(base_name_*_fold.txt) or create your own text files for the split.
- Input the preprocessed data directory path into the
- To train the Boundary Aware Point Sampling model, please modify the following four configurations in
train_configs/tgnet_bdl.py:original_data_obj_path,original_data_json_path,bdl_cache_path, andload_ckpt_path.
- After modifying the configurations, train the Boundary Aware Point Sampling model as follows.
start_train.py \ --model_name "tgnet_bdl" \ --config_path "train_configs/tgnet_bdl.py" \ --experiment_name "your_experiment_name" \ --input_data_dir_path "path/to/save/preprocessed_data" \ --train_data_split_txt_path "base_name_train_fold.txt" \ --val_data_split_txt_path "base_name_val_fold.txt"
- This is the implementation of model TSegNet. Please refer to the paper for detail.
- First, The centroid prediction module has to be trained first in tsegnet. To train the centroid prediction module, please modify the
run_tooth_seg_mentation_moduleparameter to False in thetrain_configs/tsegnet.pyfile.
- And train the centroid prediction module by entering the following command.
start_train.py \ --model_name "tsegnet" \ --config_path "train_configs/tsegnet.py" \ --experiment_name "your_experiment_name" \ --input_data_dir_path "path/to/save/preprocessed_data" \ --train_data_split_txt_path "base_name_train_fold.txt" \ --val_data_split_txt_path "base_name_val_fold.txt" - Once the training of the centroid prediction module is completed, please update the
pretrained_centroid_model_pathintrain_configs/tsegnet.pywith the checkpoint path of the trained centroid prediction module. Also, setrun_tooth_segmentation_moduleto True. - And please train the tsegnet model by entering the following command.
start_train.py \ --model_name "tsegnet" \ --config_path "train_configs/tsegnet.py" \ --experiment_name "your_experiment_name" \ --input_data_dir_path "path/to/save/preprocessed_data" \ --train_data_split_txt_path "base_name_train_fold.txt" \ --val_data_split_txt_path "base_name_val_fold.txt"
- pointnet | pointnet++ | dgcnn | pointtransformer
- This model directly applies the point cloud segmentation method to tooth segmentation.
- train models by entering the following command.
start_train.py \ --model_name "pointnet" \ --config_path "train_configs/pointnet.py" \ --experiment_name "your_experiment_name" \ --input_data_dir_path "path/to/save/preprocessed_data" \ --train_data_split_txt_path "base_name_train_fold.txt" \ --val_data_split_txt_path "base_name_val_fold.txt"
- To test the performance of the model used in the challenge, please switch to the challenge_branch (refer to the notice at the top).
- We offer six models(tsegnet | tgnet(ours) | pointnet | pointnetpp | dgcnn | pointtransformer).
- All of the checkpoint files for each model are in (https://drive.google.com/drive/folders/15oP0CZM_O_-Bir18VbSM8wRUEzoyLXby?usp=sharing). Download ckpts(new).zip and unzip all of the checkpoints.
- Inference with tsegnet / pointnet / pointnetpp / dgcnn / pointtransformer
python start_inference.py \ --input_dir_path obj/file/parent/path \ --split_txt_path base_name_test_fold.txt \ --save_path path/to/save/results \ --model_name tgnet_fps \ --checkpoint_path your/model/checkpoint/path - Inference with tgnet(ours)
python start_inference.py \ --input_dir_path obj/file/parent/path \ --split_txt_path base_name_test_fold.txt \ --save_path path/to/save/results \ --model_name tgnet_fps \ --checkpoint_path your/tgnet_fps/checkpoint/path --checkpoint_path_bdl your/tgnet_bdl/checkpoint/path- Please input the parent path of the original mesh obj files instead of the preprocessed sampling points in
--input_data_dir_pathfor training. The inference process will handle the farthest point sampling internally. - For
split_txt_path, provide the test split fold's casenames in the same format as used during training.
- Please input the parent path of the original mesh obj files instead of the preprocessed sampling points in
- Predicted results are saved in save_path like below... It has the same format as the ground truth json file.
--save_path ----00OMSZGW_lower.json ----00OMSZGW_upper.json ----0EAKT1CU_lower.json ----0EAKT1CU_upper.json and so on... - the inference config in "inference_pipelines.infenrece_pipeline_maker.py" has to be the same as the model of the train config. If you change the train config, then you have to change the inference config.
-
The checkpoints we provided were trained for 60 epochs using the train-validation split provided in the dataset drive link(
base_name_train_fold.txt,base_name_val_fold.txt). The results obtained using the test split(base_name_test_fold.txt) are as follows(IoU -> Intersection over Union(TSA in challenge) // CLS -> classification accuracy(TIR in challenge)).
-
The results may look like this.
- We provide the evaluation and visualization code.
- You can execute the following code to test on a pair of obj/gt json file:
eval_visualize_results.py \ --mesh_path path/to/obj_file \ --gt_json_path path/to/gt_json_file \ --pred_json_path path/to/predicted_json_file(a result of inference code) - With just a few modifications to the provided code, you can write code to test all the results.
To set up the environment for the ToothGroupNetwork project using Docker, follow these steps:
-
Navigate to the Repository: Open a terminal and navigate to the root directory of the ToothGroupNetwork repository:
cd path/to/ToothGroupNetwork -
Build the Docker Image: Run the following command to build the Docker image with the name
tooth_3d_env:docker build -t tooth_3d_env . -
Run the Docker Container: After the image is built, you can run the container with the following command:
docker run --gpus all -it --name tooth_3d_env tooth_3d_env
-
Access the Environment: Once the container is running, you will have access to a bash shell inside the Docker environment where you can execute your code and run experiments.
-
Stopping the Container: To stop the container, you can use:
docker stop tooth_3d_env
-
Restarting the Container: If you want to restart the container later, use:
docker start -ai tooth_3d_env
Make sure you have Docker installed and running on your machine before following these steps.
After downloading the necessary files from this Google Drive link, follow these steps to copy and unzip the files in your Docker container:
-
Copy the Downloaded Files to the Docker Container: Replace
[docker_id]with the actual ID of your running Docker container (you can find it by runningdocker ps).docker cp 'ground-truth_labels_instances.zip' [docker_id]:/workspace/assets docker cp '3D_scans_per_patient_obj_files.zip' [docker_id]:/workspace/assets docker cp 'ckpts(new).zip' [docker_id]:/workspace/assets docker cp 'base_name_test_fold.txt' [docker_id]:/workspace/assets
-
Access the Docker Container: If you are not already inside the Docker container, you can access it using:
docker exec -it [docker_id] /bin/bash -
Unzip the Files: Inside the Docker container, create the necessary directories and unzip the files:
mkdir -p /workspace/assets/ground_labels mkdir -p /workspace/assets/3D_scans mkdir -p /workspace/assets/checkpoints mkdir -p /workspace/assets/base_names # Create specific folders for unzipped contents mkdir -p /workspace/assets/ground_labels/ground-truth_labels_instances mkdir -p /workspace/assets/3D_scans/3D_scans_per_patient_obj_files mkdir -p /workspace/assets/checkpoints/ckpts_new mkdir -p /workspace/assets/base_names/base_name_test_fold # Unzip the files into their respective directories unzip /workspace/assets/ground-truth_labels_instances.zip -d /workspace/assets/ground_labels/ground-truth_labels_instances unzip /workspace/assets/3D_scans_per_patient_obj_files.zip -d /workspace/assets/3D_scans/3D_scans_per_patient_obj_files unzip /workspace/assets/ckpts(new).zip -d /workspace/assets/checkpoints/ckpts_new cp /workspace/assets/base_name_test_fold.txt /workspace/assets/base_names/base_name_test_fold/
-
Verify the Unzipped Files: You can list the contents of the directories to ensure that the files have been unzipped correctly:
ls /workspace/assets/ground_labels/ground-truth_labels_instances ls /workspace/assets/3D_scans/3D_scans_per_patient_obj_files ls /workspace/assets/checkpoints/ckpts_new ls /workspace/assets/base_names/base_name_test_fold
Make sure to replace [docker_id] with the actual ID of your running Docker container.
To run the inference on the test data, use the following command inside the Docker container:
python start_inference.py \
--input_dir_path /workspace/assets/3D_scans/3D_scans_per_patient_obj_files \
--split_txt_path /workspace/assets/base_names/base_name_test_fold.txt \
--save_path /workspace/results/test \
--model_name tgnet_fps \
--checkpoint_path /workspace/assets/checkpoints/ckpts_new/tgnet_fps.h5To evaluate the results and visualize the performance of the model, you can use the following command inside the Docker container. Make sure to replace 013FHA7K with the appropriate case name for your specific model.
python eval_visualize_results.py \
--mesh_path /workspace/assets/3D_scans/3D_scans_per_patient_obj_files/013FHA7K/013FHA7K_lower.obj \
--gt_json_path /workspace/assets/ground_labels/ground-truth_labels_instances/013FHA7K/013FHA7K_lower.json \
--pred_json_path /workspace/results/test/013FHA7K_lower.json--mesh_path: This is the path to the target model file (OBJ format). In this case, it is013FHA7K_lower.obj, which should match the name of the corresponding result.--gt_json_path: This is the path to the ground truth JSON file, which contains the expected results for the model. The file013FHA7K_lower.jsonis located in theground_labels/ground-truth_labels_instancesdirectory.--pred_json_path: This is the path to the predicted JSON file generated from the inference step. The file013FHA7K_lower.jsonis one of the results of the inference and is located in theresults/testdirectory.
Ensure that the names of the OBJ and JSON files match (e.g., 013FHA7K_lower.obj and 013FHA7K_lower.json) to avoid any mismatches during evaluation.
When you run the evaluation command, you might see output similar to the following:
Processing: 0 : /workspace/assets/3D_scans/3D_scans_per_patient_obj_files/013FHA7K/013FHA7K_lower.obj
Loaded ground truth labels from: /workspace/assets/ground_labels/ground-truth_labels_instances/013FHA7K/013FHA7K_lower.json
Loaded predicted labels from: /workspace/results/test/013FHA7K_lower.json
Calculating metrics...
IoU: 0.85, F1(TSA): 0.90, SEM_ACC(TIR): 0.88
Colored mesh saved as /workspace/gt_output.ply
2D projection saved as /workspace/gt_output.png
Colored mesh saved as /workspace/pred_output.ply
2D projection saved as /workspace/pred_output.png
Visualization completed successfully for 013FHA7K.
This log output provides insight into the evaluation process, including the loading of files, calculation of metrics, and saving of results.
You can check the generated PLY file here: View PLY Online.
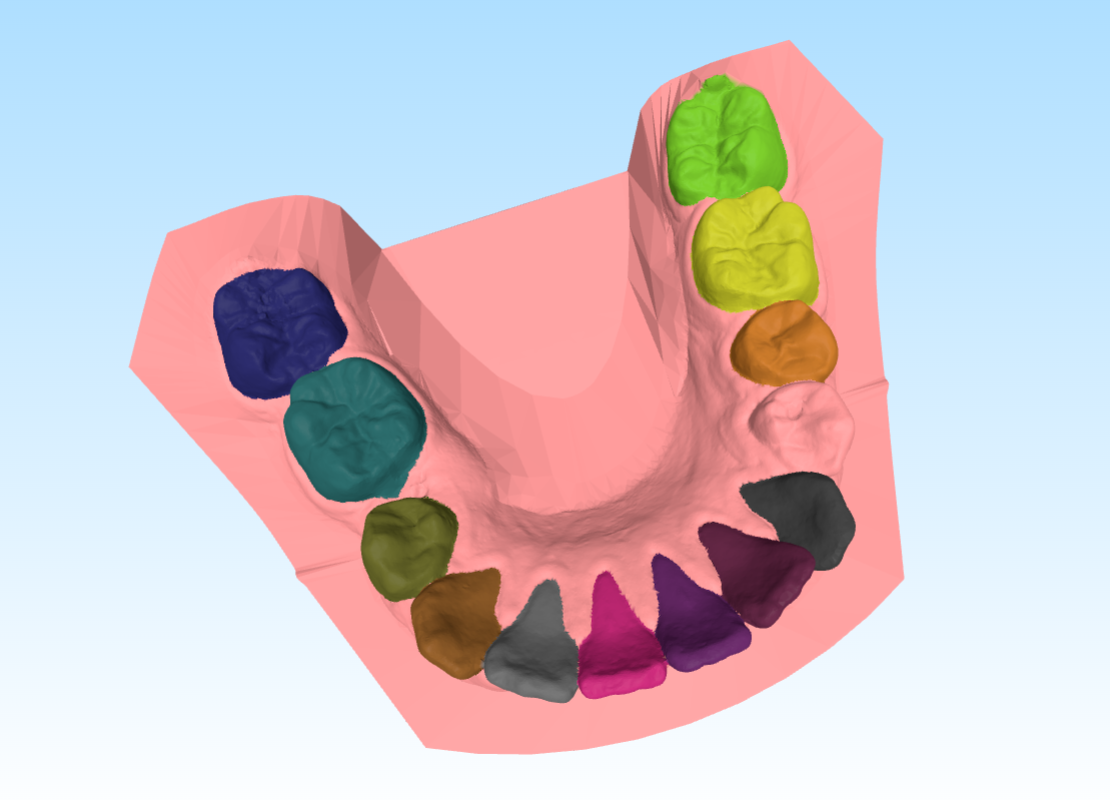
Here’s an example of the generated visualization:
- Installtion is tested on pytorch/pytorch:1.7.1-cuda11.0-cudnn8-devel(ubuntu, pytorch 1.7.1) docker image.
- It can be installed on other OS(window, etc..)
- There are some issues with RTX40XX graphic cards. plz report in issue board.
- if you have any issues while installing pointops library, please install another pointops library source in (https://github.com/POSTECH-CVLab/point-transformer).
pip install wandb pip install --ignore-installed PyYAML pip install open3d pip install multimethod pip install termcolor pip install trimesh pip install easydict cd external_libs/pointops && python setup.py install
- https://github.com/LiyaoTang/contrastBoundary.git
- https://github.com/yanx27/Pointnet_Pointnet2_pytorch
- https://github.com/POSTECH-CVLab/point-transformer.git
- https://github.com/fxia22/pointnet.pytorch
- https://github.com/WangYueFt/dgcnn