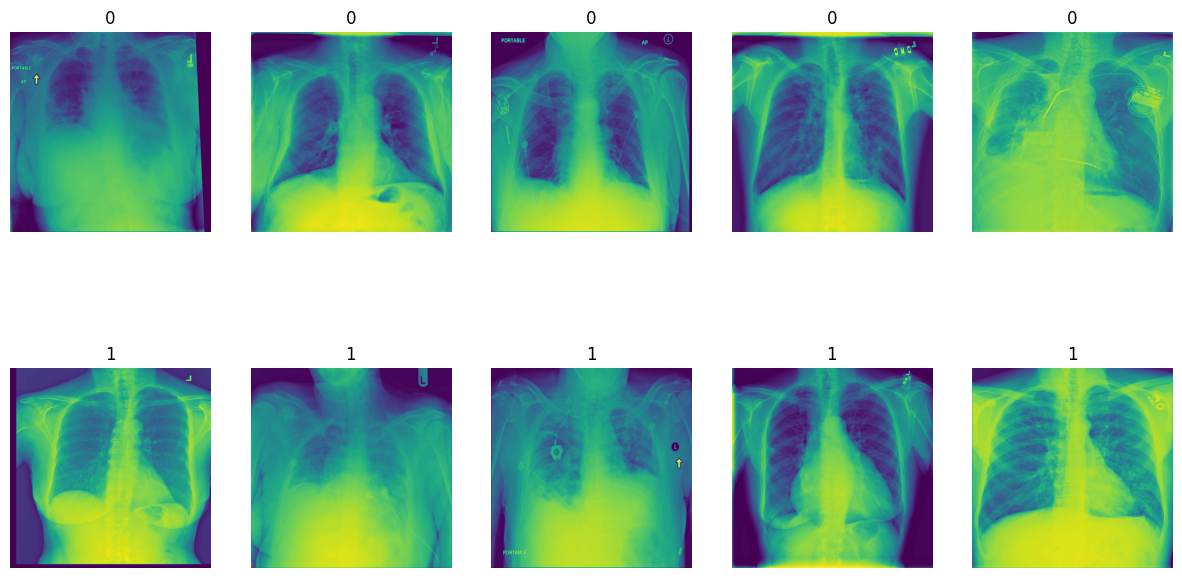
The goal of this project is to differentiate normal and abnormal chest X-ray images with high sensitivity. The dataset comprises various diseases, and the objective is to classify them into a binary task: '0' for normal and '1' for abnormal X-rays.
To achieve this, we developed a Convolutional Neural Network (CNN) capable of detecting abnormalities in X-ray images with high accuracy. We utilized CNN architectures such as VGG16, ResNet50, and InceptionV3 for feature extraction. The models were trained on 5000 images and evaluated on an additional 1000 images.
- ResNet50 outperformed VGG16 and InceptionV3.
- Augmentation did not significantly improve model performance.
- Experimented with different optimization techniques and loss functions for better results.
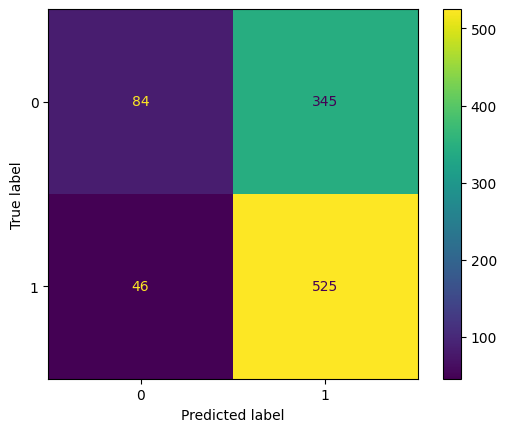
- Despite a promising recall score, initial model performance had room for improvement.
- VAL_ACCURACY is actually the model's TEST ACCURACY, it's the name only, so don't be confused by that.
| Models (12 Models) | Accuracy | Recall | Precision |
|---|---|---|---|
| val_accuracy Inc. With AUg | 0.458 | 0.269 | 0.727 |
| val_accuracy Inc. WithOut AUg | 0.446 | 0.223 | 0.752 |
| val_accuracy ResNet With AUg | 0.608 | 0.674 | 0.709 |
| val_accuracy ResNet WithOut AUg | 0.643 | 0.822 | 0.690 |
| val_accuracy VGG With AUg | 0.638 | 0.770 | 0.713 |
| val_accuracy VGG WithOut AUg | 0.618 | 0.850 | 0.645 |
| val_recall Inc. With AUg | 0.501 | 0.352 | 0.751 |
| val_recall Inc. WithOut AUg | 0.626 | 0.794 | 0.683 |
| val_recall ResNet With AUg | 0.598 | 0.540 | 0.775 |
| val_recall ResNet WithOut AUg | 0.528 | 0.389 | 0.774 |
| val_recall VGG With AUg | 0.414 | 0.153 | 0.746 |
| val_recall VGG WithOut AUg | 0.345 | 0 | 0 |
- Implemented a trick by changing the optimizer to 'adamax' and using Binary Cross Entropy as the loss function.
- Also by, instead of using threshold of 0.5 , we are going to use threshold of 0.3 to classify with high recall score.
- This simple change SKYROCKETED the Recall Score.
- Achieved high sensitivity in detecting abnormal X-ray images by using RESNET -50 as PRE-TRAINED MODEL.
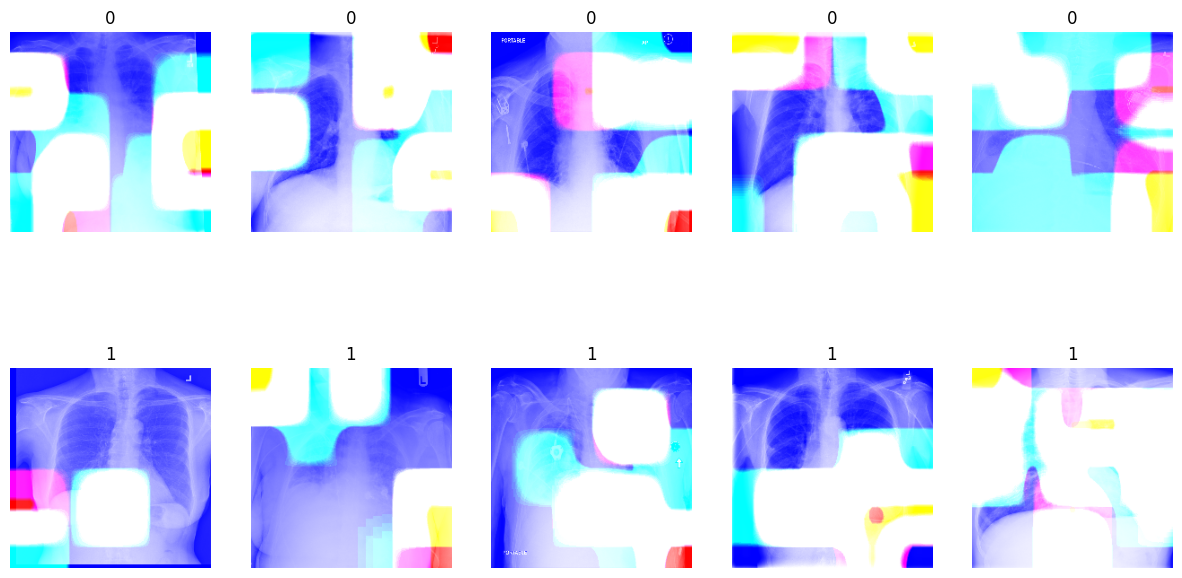
- Utilized Grad-CAMs to visualize important regions in images for classification.
- The model focuses on specific features crucial for accurate classification of lung diseases.
- Gradient-weighted Class Activation Mapping (Grad-CAM) used to understand important regions in images.
- White areas in Grad-CAMs indicate where the model focuses its attention for predictions.
- Changes in these regions impact the model's ability to classify different lung diseases accurately.
To replicate the results or explore the code further, follow these steps:
- Clone the repository to your local machine.
- Ensure you have all the dependencies installed. Check
requirements.txtfor details. - Open
main-file.ipynbusing Jupyter Notebook or any compatible environment. - Follow the instructions and run the code cells to analyze the dataset, train the model, and evaluate the results.
This project is licensed under the terms of the MIT License. Feel free to use the code and resources for educational purposes or to build upon them for your own projects.
For any inquiries or feedback, feel free to contact the author:
- Author: Luficer G
- Email: [email protected]
Start exploring and learning from this project today! If you find it useful, don't forget to give it a star ⭐️. Thank you for your interest!