This repository hosts fast parallel DBSCAN clustering code for low dimensional Euclidean space. The code automatically uses the available threads on a parallel shared-memory machine to speedup DBSCAN clustering. It stems from a paper presented in SIGMOD'20: Theoretically Efficient and Practical Parallel DBSCAN.
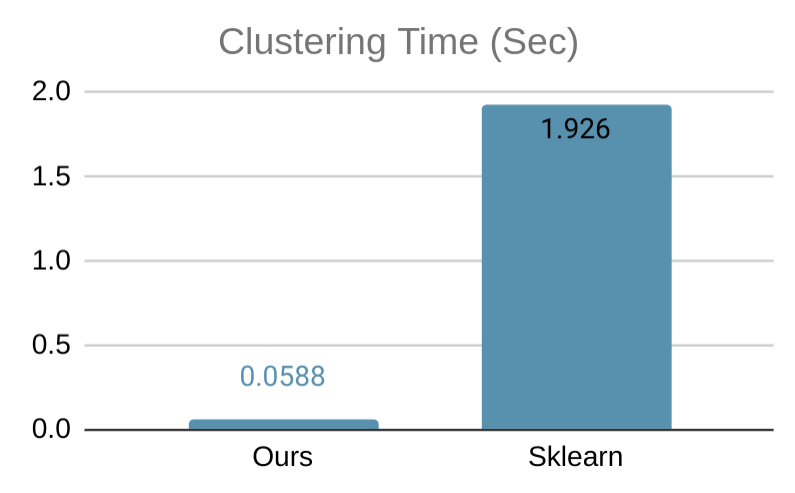
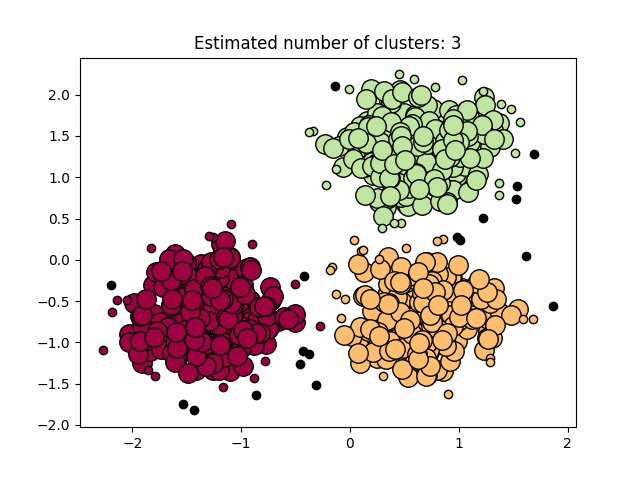
Our software on 1 thread is on par with all serial state-of-the-art DBSCAN packages, and provides additional speedup via multi-threading. Below, we show a simple benchmark comparing our code with the DBSCAN implementation of Sklearn, tested on a 6-core computer with 2-way hyperthreading using a 2-dimensional data set with 50000 data points, where both implementation uses all available threads. Our implementation is more than 32x faster. We also show a visualization of the clustering result on a smaller data set.
Data sets with dimensionality 2 - 20 are supported by default, which can be modified by modifying DBSCAN_MIN_DIMS and DBSCAN_MAX_DIMS in the source code.
There are two ways to install it:
- Install it using PyPI:
pip3 install --user dbscan(you can find the wheels here). - To build from scratch for testing:
pip3 install -e .from the project root directory.
An example for using the Python module is provided in example.py. It generates the clustering example above.
from dbscan import DBSCAN
labels, core_samples_mask = DBSCAN(X, eps=0.3, min_samples=10)
X: A 2-D Numpy array containing the input data points. The first dimension ofXis the number of data pointsn, and the second dimension is the data set dimensionality (the maximum supported dimensionality is 20).eps: The epsilon parameter (default 0.5).min_samples: The minPts parameter (default 5).
labels: A lengthnNumpy array (dtype=np.int32) containing cluster IDs of the data points, in the same ordering as the input data. Noise points are given a pseudo-ID of-1.core_samples_mask: A lengthnNumpy array (dtype=np.bool) masking the core points, in the same ordering as the input data.
We provide a complete example below that generates a toy data set, computes the DBSCAN clustering, and visualizes the result as shown in the plot above.
import numpy as np
from sklearn.datasets import make_blobs
from sklearn.preprocessing import StandardScaler
# #############################################################################
# Generate sample data
centers = [[1, 1], [-1, -1], [1, -1]]
X, labels_true = make_blobs(n_samples=750, centers=centers, cluster_std=0.4,
random_state=0)
X = StandardScaler().fit_transform(X)
# #############################################################################
# Compute DBSCAN
# direct call of the C API:
from dbscan import DBSCAN
labels, core_samples_mask = DBSCAN(X, eps=0.3, min_samples=10)
# OR calling our sklearn API:
# from dbscan import sklDBSCAN as DBSCAN
# db = DBSCAN(eps=0.3, min_samples=10).fit(X)
# core_samples_mask = np.zeros_like(db.labels_, dtype=bool)
# core_samples_mask[db.core_sample_indices_] = True
# labels = db.labels_
# #############################################################################
# Plot result
import matplotlib.pyplot as plt
n_clusters_ = len(set(labels)) - (1 if -1 in labels else 0)
n_noise_ = list(labels).count(-1)
# Black removed and is used for noise instead.
unique_labels = set(labels)
colors = [plt.cm.Spectral(each)
for each in np.linspace(0, 1, len(unique_labels))]
for k, col in zip(unique_labels, colors):
if k == -1:
# Black used for noise.
col = [0, 0, 0, 1]
class_member_mask = (labels == k)
xy = X[class_member_mask & core_samples_mask]
plt.plot(xy[:, 0], xy[:, 1], 'o', markerfacecolor=tuple(col),
markeredgecolor='k', markersize=14)
xy = X[class_member_mask & ~core_samples_mask]
plt.plot(xy[:, 0], xy[:, 1], 'o', markerfacecolor=tuple(col),
markeredgecolor='k', markersize=6)
plt.title('Estimated number of clusters: %d' % n_clusters_)
plt.show()
Compile and run the program:
mkdir build
cd build
cmake ..
cd executable
make -j # this will take a while
./dbscan -eps 0.1 -minpts 10 -o clusters.txt <data-file>
The <data-file> can be any CSV-like point data file, where each line contains a data point -- see an example here. The data file can be either with or without header. The cluster output clusters.txt will contain a cluster ID on each line (other than the first-line header), giving a cluster assignment in the same ordering as the input file. A noise point will have a cluster ID of -1.
Create your own caller header and source file by instantiating the DBSCAN template function in "dbscan/algo.h".
dbscan.h:
template<int dim>
int DBSCAN(int n, double* PF, double epsilon, int minPts, bool* coreFlagOut, int* coreFlag, int* cluster);
// equivalent to
// int DBSCAN(intT n, floatT PF[n][dim], double epsilon, intT minPts, bool coreFlagOut[n], intT coreFlag[n], intT cluster[n])
// if C++ syntax was a little more flexible
template<>
int DBSCAN<3>(int n, double* PF, double epsilon, int minPts, bool* coreFlagOut, int* coreFlag, int* cluster);dbscan.cpp:
#include "dbscan/algo.h"
#include "dbscan.h"Calling the instantiated function:
int n = ...; // number of data points
double data[n][3] = ...; // data points
int labels[n]; // label ids get saved here
bool core_samples[n]; // a flag determining whether or not the sample is a core sample is saved here
{
int ignore[n];
DBSCAN<3>(n, (void*)data, 70, 100, core_samples, ignore, labels);
}Doing this will only compile the function for the number of dimensions that you want, which saves on compilation time.
You can also include the "dbscan/capi.h" and define your own DBSCAN_MIN_DIMS and DBSCAN_MAX_DIMS macros the same way the Python extension uses it. The function exported has the following signature.
extern "C" int DBSCAN(int dim, int n, double* PF, double epsilon, int minPts, bool* coreFlag, int* cluster);Right now, the only two files that are guaranteed to remain in the C/C++ API are "dbscan/algo.h" and "dbscan/capi.h" and the functions named DBSCAN within.
If you use our work in a publication, we would appreciate citations:
@inproceedings{wang2020theoretically,
author = {Wang, Yiqiu and Gu, Yan and Shun, Julian},
title = {Theoretically-Efficient and Practical Parallel DBSCAN},
year = {2020},
isbn = {9781450367356},
publisher = {Association for Computing Machinery},
address = {New York, NY, USA},
url = {https://doi.org/10.1145/3318464.3380582},
doi = {10.1145/3318464.3380582},
booktitle = {Proceedings of the 2020 ACM SIGMOD International Conference on Management of Data},
pages = {2555–2571},
numpages = {17},
keywords = {parallel algorithms, spatial clustering, DBScan},
location = {Portland, OR, USA},
series = {SIGMOD ’20}
}