Qin Zhu, Kim Lab, University of Pennsylvania
This is a tool for interactive visualization of C. elegans embryogenesis single cell data published with Packer, J.S., Zhu, Q., et al., 2019.
It has entire C. elegans embryogensis data built in, with features specifically designed for showing analysis results in the C. elegans paper, such as co-visualization of umap, lineage annotation and lineage tree.
For using VisCello for other single cell data visualization, please check https://github.com/qinzhu/VisCello.
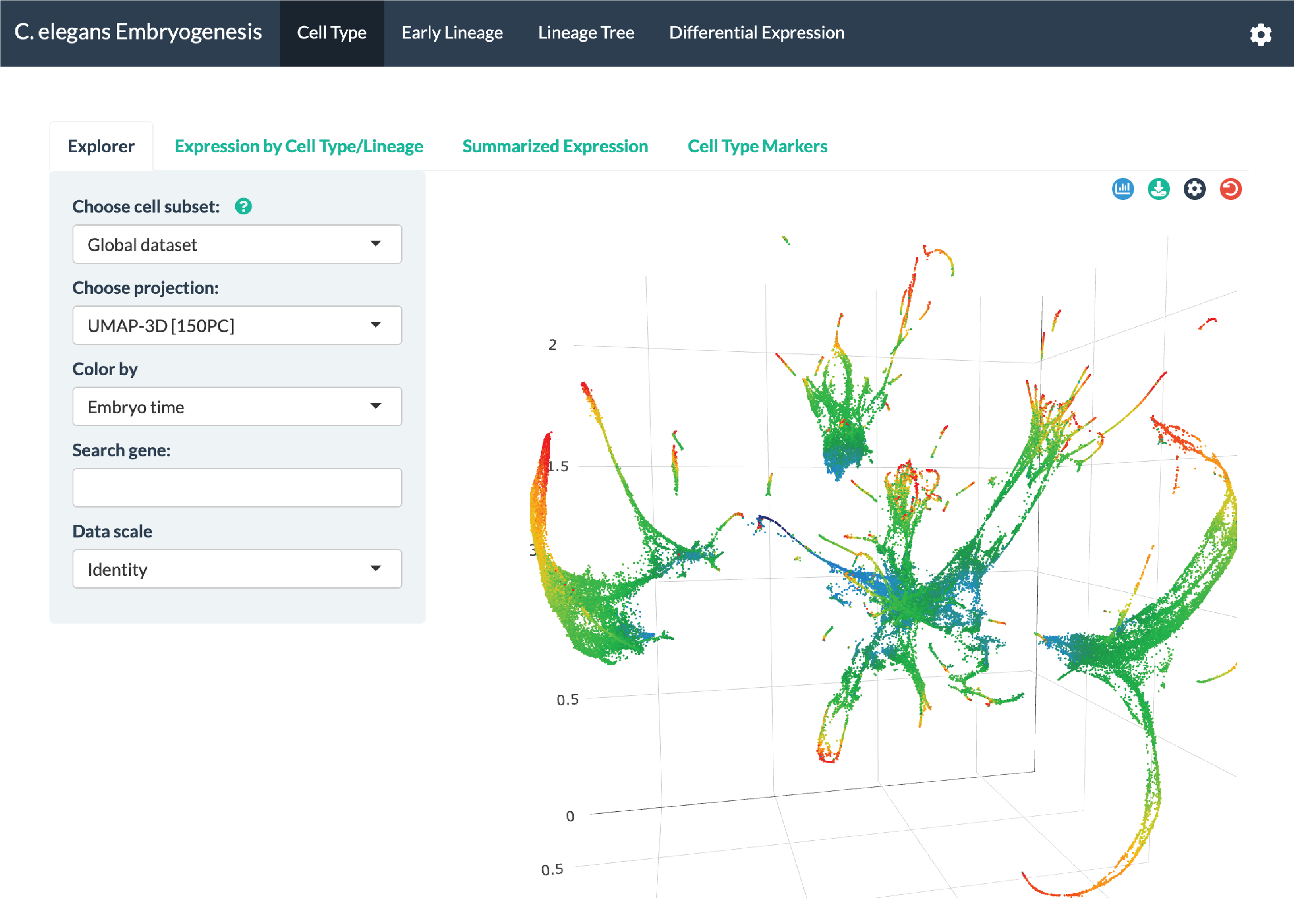
Screenshot:
Link: https://cello.shinyapps.io/celegans/
Bugs you found with the online tool please post to this github repo.
-
C. elegans L2 data: https://github.com/qinzhu/Celegans.L2.Cello
-
C. elegans Tintori et al. data (up to 16 cell stage): https://github.com/qinzhu/Celegans.Tintori.Cello
Due to large data file currently being hosted on Git LFS, you cannot use devtools::install_github to install this package. Please follow protocol listed below to install:
-
Windows and Linux: install git from https://git-scm.com/book/en/v2/Getting-Started-Installing-Git.
MACOS and Linux: Install git-lfs from https://git-lfs.github.com/ (See FAQ for example installation using brew or conda).
Any system: Install R (>3.5.0 required) and latest bioconductor (code below, copy and paste inside R):
if (!requireNamespace("BiocManager")) install.packages("BiocManager") BiocManager::install() # For first time installer, if prompted "Update all/some/none? [a/s/n]", type a and press return. -
In terminal copy paste followng line by line with return:
git lfs clone https://github.com/qinzhu/VisCello.celegans.git R #Launch R, Windows user open R/Rstudio, setwd() to parent folder of VisCello. -
Now inside R and do the following (line by line):
install.packages("devtools") devtools::install_local("VisCello.celegans", force=T) # Temporarily needed, install a version of "tidytree" package to avoid a bug in newer version packageurl <- "https://cran.r-project.org/src/contrib/Archive/tidytree/tidytree_0.2.4.tar.gz" install.packages(packageurl, repos=NULL, type="source")
Now VisCello is ready to go! To launch Viscello, in R:
library(VisCello.celegans) cello()
If you have also installed VisCello, please make sure you do not load VisCello in the same session, as some functions are re-used and may lead to conflicts.
-
Q: Got installation error:
Error in readRDS("data/eset.rds"): unknown input formatA: Install git-lfs from Install git-lfs from https://git-lfs.github.com/ and then go to step 1. Example code using brew or conda:
brew install git-lfs git lfs install git-lfs clone https://github.com/qinzhu/VisCello.celegans.gitconda install -c conda-forge git-lfs git lfs install git-lfs clone https://github.com/qinzhu/VisCello.celegans.git -
Q: Got installation error when running devtools::install_local:
package 'AnnotationDbi' was built under R version xxx, lazy loading failed for package xx.A: Run following code in R:
BiocManager::install(c("celegans.db", "GO.db", "DO.db")) install.packages("VisCello.celegans", repos = NULL, type = "source") -
Q: Can I use VisCello to explore my own data?
A: Yes. Please use the general version: https://github.com/qinzhu/VisCello and kindly cite VisCello if you use it for publication.
Packer, J. S., Q. Zhu, C. Huynh, P. Sivaramakrishnan, E. Preston, H. Dueck, D. Stefanik, K. Tan, C. Trapnell, J. Kim, R. H. Waterston and J. I. Murray (2019). A lineage-resolved molecular atlas of C. elegans embryogenesis at single-cell resolution. Science: eaax1971.
Q. Zhu, J. I. Murray, K. Tan, J. Kim, qinzhu/VisCello: VisCello v1.0.0 (2019; https://zenodo.org/record/3262313).